Figure 2.
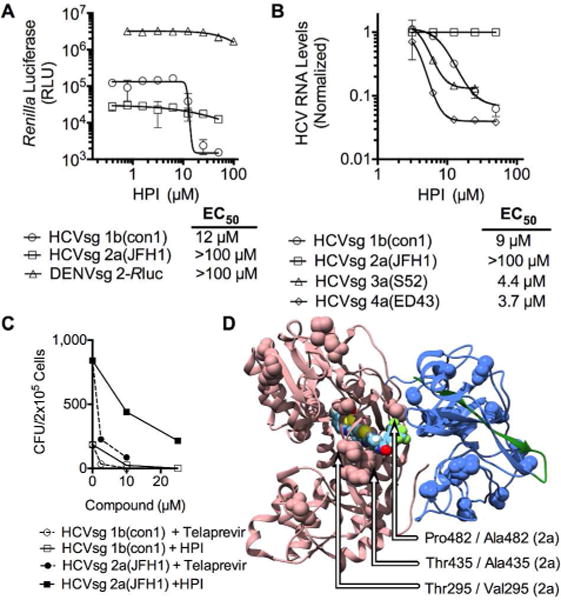
HPI specificity. (A) The ability of HPI to reduce cellular content of Renilla luciferase tagged subgenomic replicons made from HCV genotype 1b (HCVsg 1b(con1), circles), HCV genotype 2a (HCVsg 2a(JFH1), squares) and dengue virus strain 2 (DENVsg 2, triangles) (B) Effect of various HPI concentrations on relative levels of subgenomic replicon RNA, as measured by quantitative reverse-transcriptase PCR, with data normalized to RNA levels seen in cells treated with DMSO only. (C) Colony formation units (CFU) of Huh7.5 cell cultures harboring the HCVsg 1b(con1) or the HCVsg 2a(JFH1) replicon. Cells were initially plated at 2 × 105 cells/dish, and G418-resistant colonies were stained with crystal violet after 3 weeks of antibiotic selection. Note CFUs for the HCVsg 2a(JFH1) replicon were about 10 times higher than CFUs observed with HCVsg 1b(con1) in the absence of HPI or telaprevir. (D) Unique residues in genotype 2a(JFH1) are highlighted on the scNS4A-NS3 structure in which HPI is docked. Residues pesent in 2a(JFH1) NS3 but not genotypes 1a(H77), 1b(con1), 3a(S52), or 4a(ED42) are highlighted as spheres with unique amino acids within 5 Å of HPI noted with arrows. Sequence alignments are shown in Figure S1 (Supporting Information).
