Figure 5.
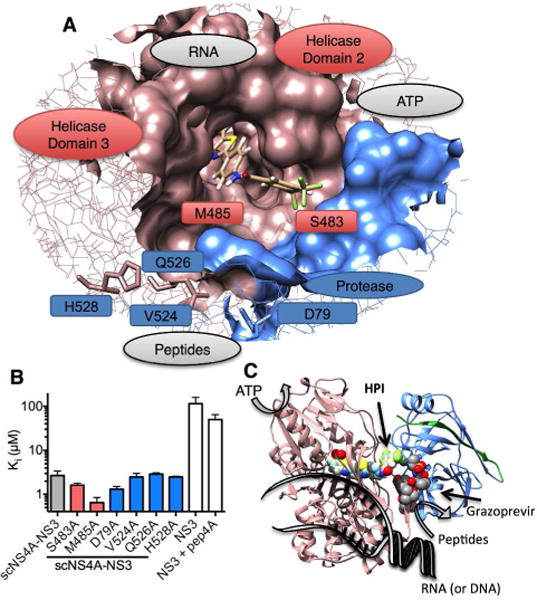
The possible HPI-binding site on NS3. (A) Position of HPI when docked in the full-length HCV NS3 structure (PDB 1CU1). The putative HPI binding site is highlighted as a surface on a wireframe NS3 model with the helicase red and protease blue. Residues targeted for site-directed mutagenesis are shown as sticks. Natural ligand-binding sites are labeled in grey. (B) Inhibitory constant (Ki) describing the ability of HPI to inhibit NS3-catalyzed peptide hydrolysis. Wildtype scNS4A-NS3 is grey, amino acid substitutions in the putative HPI-binding site are red, and amino acid substitutions in the cleft that binds protease substrates are blue. Full-length NS3 constructs lacking the covalent tether to NS4A are white. (C) Position of docked HPI in realtion to the various NS3 active sites, ligand binding clefts, and the peptidomimetic protease inhibitor-binding site. Position of grazoprevir was determined by aligning the grazoprevir costructure with the NS3 protease (PDB file 3SUD)31 with the structure of full-length NS3 (PDB file 1CU1).26 The protease domain is blue, NS4A is green, and the helicase is red. Models were rendered using UCSF Chimera 1.8.34
