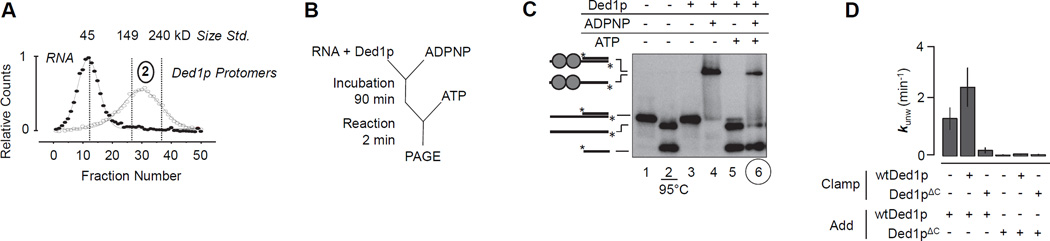
Figure 3. Unwinding involves distinct loading and unwinding protomers.
(A) Fractionation of of RNA (16 bp, filled circles) and RNA with Ded1p and ADPNP (open circles) on a sucrose gradient (6–40%). Curves represent best fits to a Gaussian distribution. Dashed lines indicate peak fraction numbers of protein markers sedimented under identical conditions.
(B) Reaction scheme for testing RNA unwinding with pre-bound (clamped) Ded1p protomers.
(C) Representative non-denaturing PAGE for RNA unwinding with pre-bound (clamped) Ded1p protomers, and associated controls (0.6 µM Ded1p; 0.5 nM RNA, both strands radiolabeled, 50 µM ADPNP/Mg2+, where applicable; and 2 mM ATP/Mg2+, where applicable). To samples in lanes 4 – 6, 1 µM 73 ntRNA scavenger was added after the reaction to remove loosely associated Ded1p from the labeled RNA.
(D) Impact of pre-bound (clamped) Ded1p or Ded1pΔC on the unwinding rate constant kunwmax. See also Figure S3.