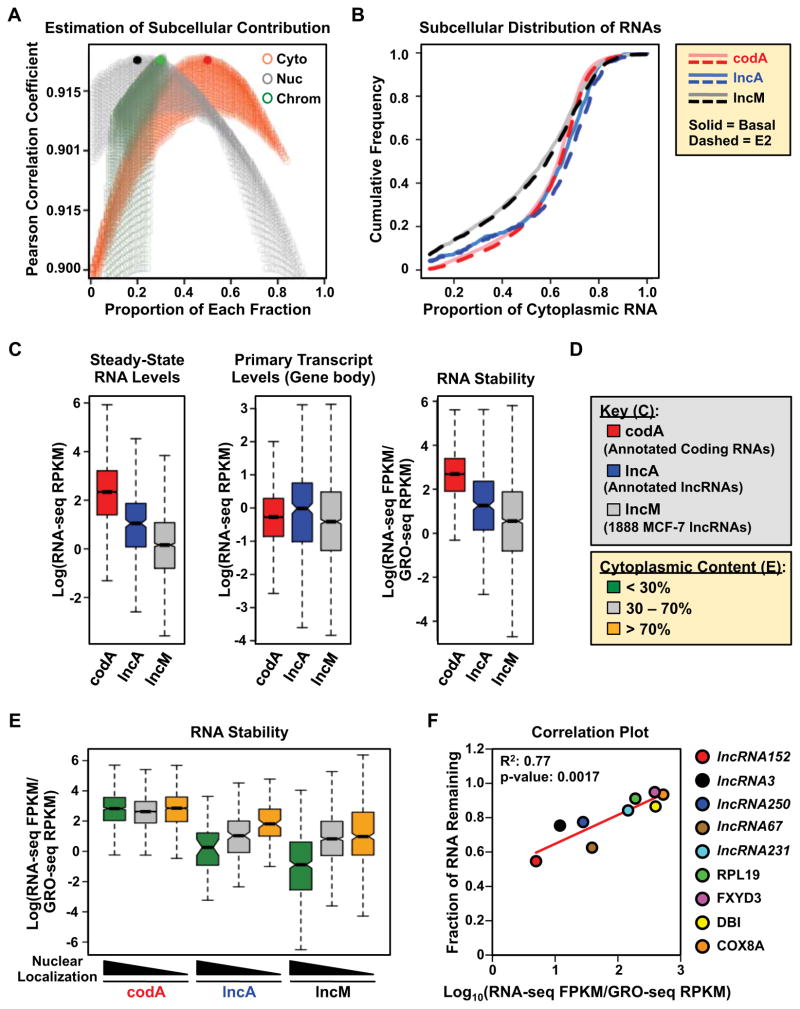
Figure 2. Nucleus-enriched lncRNAs are less stable than cytoplasm-enriched lncRNAs.
(A) Estimation of the contribution of each subcellular fraction to the total RNA content, based on the relationship a x Cyto +b x Nuc +c x Chr = Total. Pearson correlation coefficients are plotted for every pair of a x Cyto +b x Nuc +c x Chr and Total as the contribution of each fraction, a, b and c, are sampled from 0.01 to 0.99. The combination, a = 0.5; b = 0.2; c = 0.3, gives the highest correlation (solid circles).
(B) Cumulative frequency curves showing the extent of cytoplasmic localization of codA, lncA, and lncM RNAs in basal (solid) and E2-treated (dotted) conditions.
(C) Steady-state RNA levels (from RNA-seq) (left), Primary transcript levels (from GRO-seq) (middle), and relative stability (right) of annotated mRNAs (codA), annotated lncRNAs (lncA), and the lncM lncRNA set. The relative stability of RNAs is represented by the ratio of steady-state RNA levels to nascent transcript levels.
(D) Color keys for (C) through (E).
(E) Box plot showing the relative stability of codA, lncA and lncM RNAs in untreated MCF-7 cells, grouped by the extent of cytoplasmic localization.
(F) Correlation between computed RNA stability (as in panel C, right) and experimentally determined RNA stability from actinomycin D treated MCF-7 cells. Pearson correlation coefficient.
In (C) and (E), Log = the natural log (loge).
See also Figure S2.