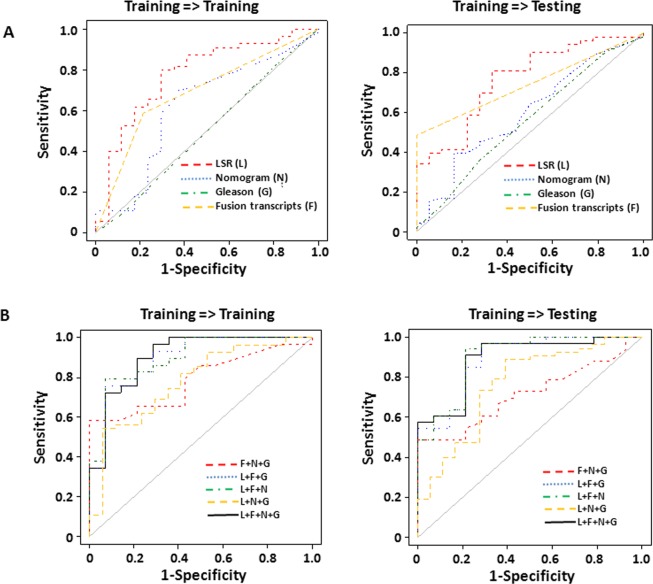
Fig 3. LSR of genome CNV from leukocytes to predict prostate cancer recurrence.
(A) LSR derived from leukocyte genome CNV predicts prostate cancer recurrence. Receiver operating curve (ROC) analyses using LSRs derived from leukocyte CNVs as prediction parameter (red) to predict prostate cancer recurrence, versus Nomogram (blue), Gleason’s grade (green) and the status of 8 fusion transcripts[14] (yellow). The samples were equally split randomly into training and testing sets 10 times. The ROC analysis represents the results from the most representative split. (B) Combination of LSR (L), Gleason’s grade (G), Nomogram (N) and the status of fusion transcripts (F) to predict prostate cancer recurrence. ROC analysis of a model combining LSR, fusion transcripts, Nomogram and Gleason’s grade using LDA is indicated by black. ROC analysis of a model combining fusion transcripts, Nomogram and Gleason’s grade using LDA is indicated by red. ROC analysis of a model combining LSR, fusion transcripts and Gleason’s grade using LDA is indicated by blue. ROC analysis of a model combining LSR, fusion transcripts and Nomogram using LDA is indicated by green. ROC analysis of a model combining LSR, Nomogram and Gleason’s grade is indicated by yellow. Similar random splits of training and testing data sets were performed as of (A).