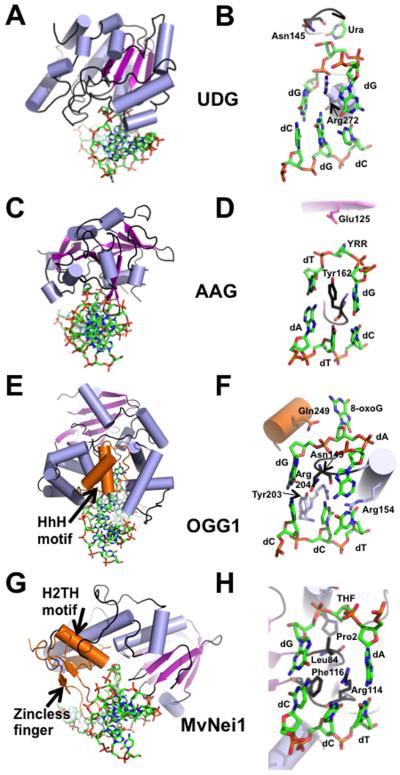
Figure 3.
Representative crystal structures from each of the 4 families of DNA glycosylases. (A) Overall DNA-bound structure of UDG bound to DNA (PDB ID: 4SKN [Slupphaug et al., 1996]). (B) Close-up view of the active site of UDG where the active site nucleophile Asp145 is mutated to Asn, Leu272 is mutated to Arg, and the cleaved uracil (Ura) remains bound in the active site pocket. (C) Human AAG (PDB ID: 1BNK [Lau et al., 1998]) bound to DNA. (D) Active site view of the AAG-DNA complex indicating an abasic pyrrolidine nucleotide (YRR) that is extruded into the active site, active site nucleophile Glu125, and Tyr162 that causes a severe kink in the DNA. (E) Overall structure of human OGG1 bound to 8-oxoG containing DNA (PDB ID: 1EBM [Bruner et al., 2000]), an example of the HhH glycosylase family. (F) Close-up view of the active site residues of OGG1 emphasizes the extrahelical 8-oxoG lesion, the active site Lys249 mutated to Gln, and the four residues that contact the estranged dC. (G) Overall structure of the viral ortholog of human NEIL1, MvNei1, bound to an abasic site analog (THF), representing the Fpg/Nei family (PDB ID: 3A46 [Imamura et al., 2009]). (H) Zoomed-in view of the MvNei1-THF complex depicting the three void-filling residues and the active site nucleophile, Pro2. For all the structures, the DNA is shown in green as a stick model, and colored by element; the α-helices are colored in light blue; β-strands are purple; loops are shown in black. The HhH motif (OGG1), the H2TH motif (MvNei1) and the zincless finger motif (MvNei1) are highlighted in orange.