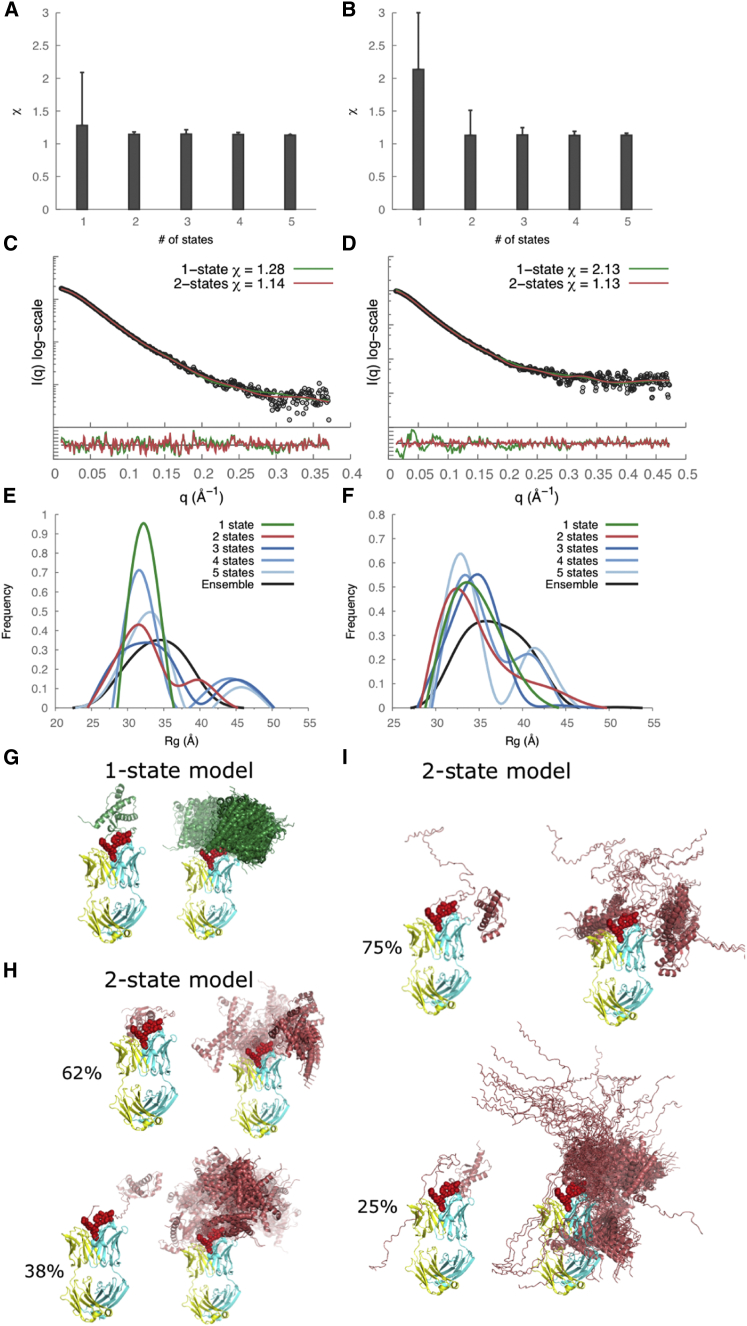
Figure 5.
SAXS data analysis and modeling for the recPrP(89–230)-Fab-P (left column) and recPrP(23–230)-Fab-P complexes (right column). (A and B) The lowest χ scores for each of the N-state models (N = 1–5) are shown with error bars indicating the range of χ values for the top 1000 models. (C and D) Comparison of the experimental SAXS profiles (black) with the computed SAXS profiles for the top-scoring single-state (green) and two-state (red) models. The lower plots show the residuals defined as (Iexp(q) − Icalc(q))/σexp(q), corresponding to the difference between the experimental and computed intensities weighted by the experimental uncertainty. (E and F) Rg distributions for the entire ensemble of 10,000 conformations and the top 1000 N-state models are shown. (G–I) Conformations of the top-scoring single-state (G) and two-state models (H and I) are shown on the left and the top 100 models aligned on the Fab structure on the right. The heavy chain of Fab is shown in cyan, the light chain in yellow, and the epitope residues in red spacefill. To see this figure in color, go online.