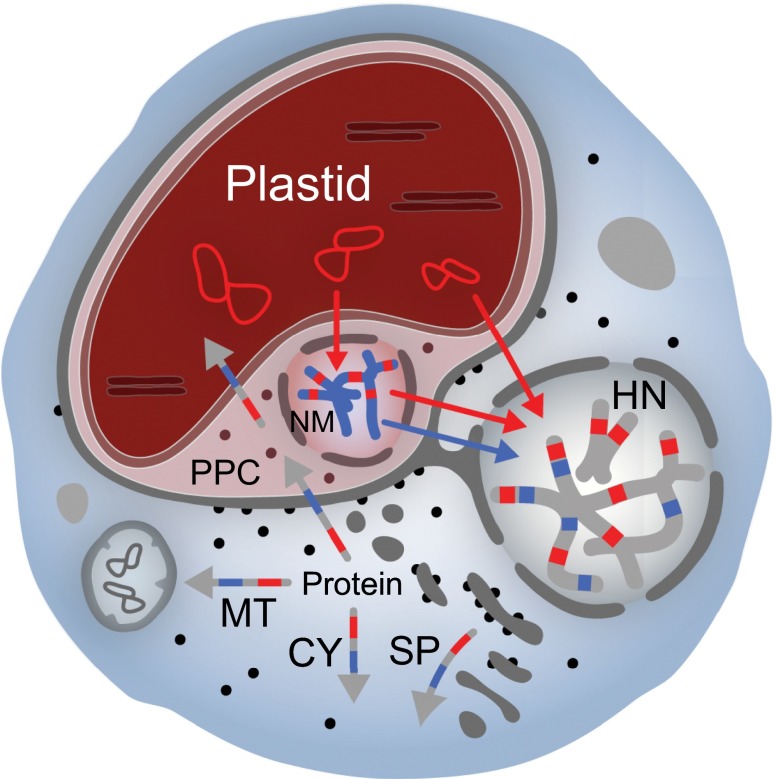
Fig. 1.
Genome and proteome mosaicism in complex algae. Diagram depicts a generic complex alga with a four-membrane secondary plastid and a nucleomorph (NM). When present, the nucleomorph resides within the periplastidial compartment (PPC), which corresponds to the cytosol of the engulfed primary algal cell. Sequenced nucleomorph genomes of cryptophyte and chlorarachniophyte algae possess at most ∼500 protein genes; most of the proteins needed to sustain the plastid and associated compartments are the products of genes residing in the host nucleus (HN). The host cell’s endomembrane system is involved in targeting nucleus-encoded proteins to the organelle. In some (but not all) complex algae, the outermost plastid membrane is physically continuous with the rough ER, allowing cotranslational insertion of organelle-targeted proteins. Blue and red arrows show endosymbiotic gene transfer events from the plastid to the primary and secondary host nuclei: their genomes are evolutionary mosaics. Multicolored arrows show possible destinations for nucleus-encoded proteins whose genes trace back to the plastid (red), nucleomorph (blue), and secondary host nucleus (gray). Evidence for all of these scenarios has come from comparative genomic investigations of several independently evolved complex algae (see text). Gene transfers involving mitochondrial DNA have been omitted for simplicity. CY, cytosol; MT, mitochondrion; SP, secretory pathway.