Fig. S1.
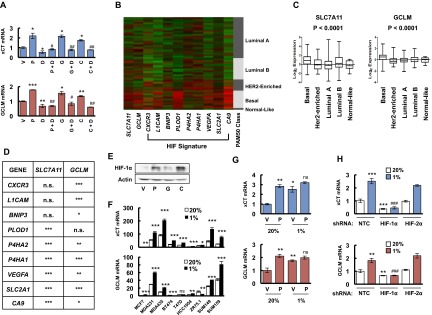
Chemotherapy and hypoxia induce xCT and GCLM expression in a HIF-1–dependent manner. (A) SUM-149 cells were treated with vehicle (V), 5 nM paclitaxel (P), 10 nM gemcitabine (G), 100 μM carboplatin (C), either alone or in combination with 100 nM digoxin (P + D, G + D, C + D) for 72 h, and RT-qPCR were performed to analyze the expression of xCT and GCLM mRNA. The results were normalized to cells treated with vehicle (mean ± SEM; n = 3). *P < 0.05, **P < 0.01, ***P < 0.001 vs. vehicle; #P < 0.05, ##P < 0.01 vs. chemotherapy alone. (B) The relative expression levels of nine mRNAs encoded by HIF target genes, as well as SLC7A11 (xCT) and GCLM mRNA, are shown for breast cancers accessed from the Cancer Genome Atlas (TCGA) database, which were classified based on the expression of a 50-mRNA (PAM50) signature that defines five molecular subtypes (Luminal A, Luminal B, HER2-enriched, Basal, and Normal-like) (22). Color code: green, less than the median; red, greater than the median. (C) Log2 expression of SLC7A11 (xCT) and GCLM mRNA from 1,215 human breast cancer specimens from TCGA database were compared among five molecular subgroups. Data were analyzed by using one-way ANOVA. (D) SLC7A11 (xCT) and GCLM mRNA levels in each of 1,215 human breast cancers from the TCGA database were compared with the levels of nine HIF-regulated mRNAs by using Pearson’s correlation test. *P < 0.05, **P < 0.01, ***P < 0.001; n.s., not significant. (E) MDA-MB-231 cells were treated with vehicle (V), 10 nM paclitaxel (P), 10 nM gemcitabine (G), or 100 μM carboplatin (C) for 72 h, and immunoblot assays were performed. (F) Nine breast cancer cell lines were exposed to 20% or 1% O2 for 24 h, total cellular RNA was isolated, and RT-qPCR was performed to analyze the expression of xCT and GCLM mRNA. The results were normalized to MCF-7 cells that were exposed to 20% O2 (mean ± SEM; n = 3). *P < 0.05, **P < 0.01, ***P < 0.001 vs. 20% O2. (G) MDA-MB-231 cells were treated with vehicle (V) or 10 nM paclitaxel (P) in 20% or 1% O2 for 72 h, total RNA was isolated, and RT-qPCR was performed to analyze the expression of xCT and GCLM mRNA (mean ± SEM; n = 3). *P < 0.05, **P < 0.01 vs. vehicle in 20% O2; ns, not significant vs. paclitaxel in 20% O2. (H) MDA-MB-231 subclones, which were stably transfected with nontargeting control shRNA (NTC) vector or expression vector encoding shRNA against HIF-1α or HIF-2α, were exposed to 20% or 1% O2 for 24 h, total RNA was isolated, and RT-qPCR was performed to analyze the expression of xCT and GCLM mRNA (mean ± SEM; n = 3). **P < 0.01, ***P < 0.001 vs. NTC in 20% O2; ###P < 0.001 vs. NTC in 1% O2.
