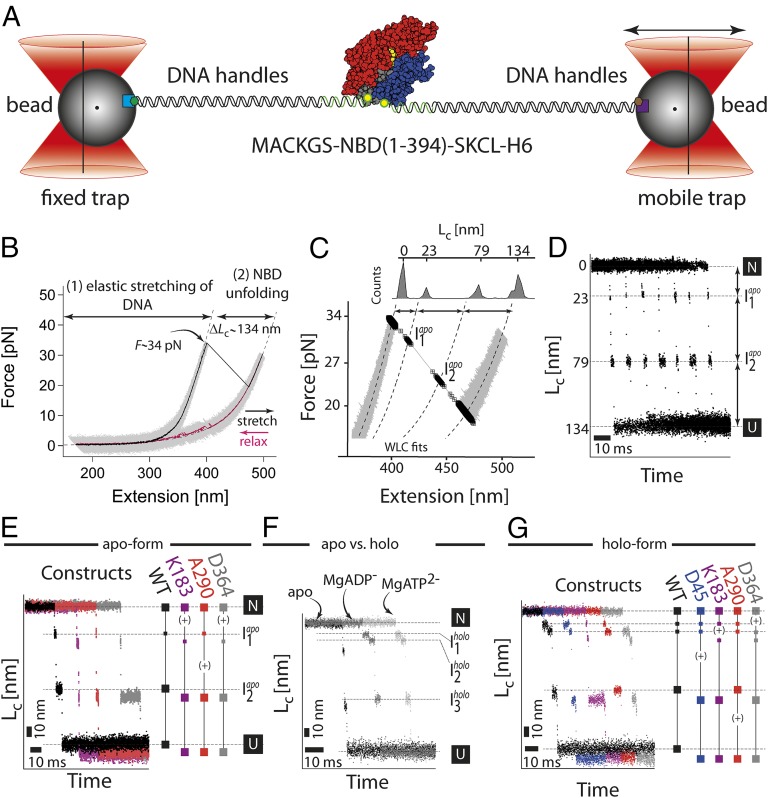
Fig. 2.
Single molecule force experiments of the NBD by optical tweezers. (A) Optical trap assay. The protein is tethered to the beads by two DNA handles containing a corresponding epitope (biotin, green hexagon; digoxygenin, brown hexagon). The connection between the DNA and protein is realized by the modification of the two cysteine residues of the protein by the single-stranded DNA–maleimide oligonucleotide complementary to the DNA handle overhang. The 1 μm-sized functionalized (α-digoxygenin, purple square; streptavidin, light blue square) glass beads are trapped in the highly focused laser beam. One of the beams is reflected by the steerable mirror, which enables pulling and stretching of a single protein. (B) Force–extension curves of a single NBD domain (the apo-form). The protein–DNA construct was stretched (black curve, 1 kHz filtered) and relaxed (purple curve, 1 kHz filtered) at a constant velocity of 20 nm/s. The trace depicts two parts corresponding to the stretching of the DNA handles and a sudden rip at 34 pN corresponding to the unfolding of the protein. The dashed lines correspond to a WLC fit to the data yielding the contour-length increase of 134 nm. (C) A magnification of the force–extension trace. The unfolding phase of the protein reveals transient populations of the intermediates (black squares; for details, see Single Molecule Force Experiments on the NBD of the DnaK Chaperone). (D) Contour-length transformation of the force–extension data. Shown are multiple unfolding phases. (E) Contour-length increase versus time for the wild-type (black) and insert variants (purple for K183-Insert, red for A290-Insert, and gray for D364-Insert). The insert variants have an additional increase in the contour length (∼7 nm) due to insertion of a highly flexible 20 aa large loop. The plus sign (+) indicates an increase in the length at a particular position. (F) Comparison of the unfolding phases between different protein forms: apo, MgATP, and MgADP. The conditions were 50 mM Tris⋅HCl, 150 mM KCl, 5 mM MgCl2, and 1 mM ATP/ADP, pH 7.8. (G) Contour-length increase versus time for the wild-type (black) and insert variants (purple for K183-Insert, red for A290-Insert, and gray for D364-Insert) under holo-conditions (1 mM ATP, 5 mM MgCl2).