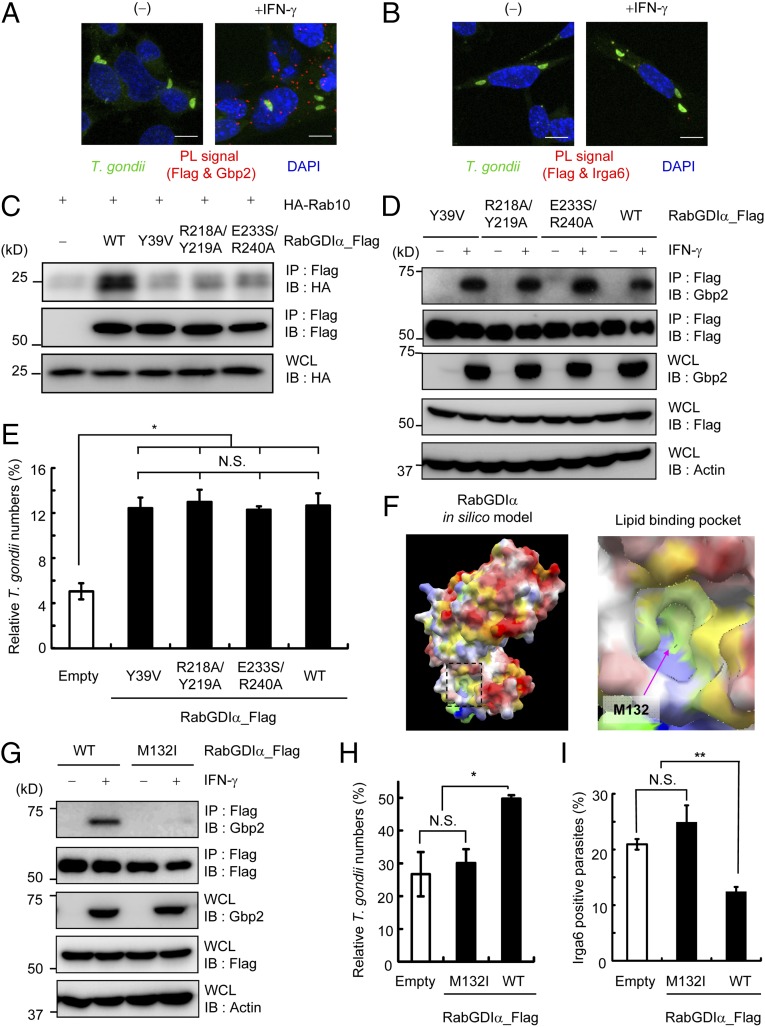
Fig. 5.
RabGDIα suppresses Gbp2-dependent responses through the lipid-binding pocket. (A and B) PL assay of Flag-tagged RabGDIα and endogenous Gbp2 (A) or Irga6 (B) in IFN-γ–stimulated or unstimulated MEFs. Cells were infected with ME49 T. gondii (moi = 4), fixed at 3 h postinfection; red, PL-positive (PL+) signals; green, T. gondii; blue, nuclei. (C) Lysates of 293T cells transiently cotransfected with indicated Flag-tagged RabGDIα vectors and/or HA-tagged Rab10 were immunoprecipitated with anti-Flag and detected using the indicated Abs by Western blot. (D) RabGDIα-deficient MEFs expressing indicated Flag-tagged RabGDIα were untreated or treated with IFN-γ and lysed. The lysates were immunoprecipitated with anti-Flag and detected using with the indicated Abs by Western blot. (E and H) T. gondii numbers at 36 h postinfection in RabGDIα-deficient MEFs stably transfected with empty (control) or indicated Flag-tagged RabGDIα expression plasmids untreated or treated with IFN-γ were analyzed by luciferase assay. Percentages of parasite numbers (calculated by luciferase counts) in IFN-γ–stimulated control or indicated RabGDIα variants-expressing cells relative to those in the unstimulated respective cells are shown as “Relative T. gondii numbers.” Indicated values are means ± SD of triplicates. (F) In silico profiling of the molecular surface of RabGDIα, colored by electrostatic potential, with red (or blue) representing the most negative (or positive) values, and by hydrophobicity, with yellow representing the most hydrophobic. The lipid-binding pocket is framed by a dashed line, and its close-up view is shown Right. The approximate location of the methionine residue 132 is indicated by an arrow. (G) Lysates of RabGDIα-deficient MEFs stably transfected with empty or indicated Flag-tagged RabGDIα expression plasmids were detected with indicated Abs by Western blot. (I) The percentage of parasites positive for Irga6 staining in RabGDIα-deficient MEFs expressing indicated RabGDIα variants at 3 h postinfection. Indicated values are means ± SD of triplicates. N.S., not significant; *P < 0.05, **P < 0.01. Data are representative of two (A–D and G) or three (E and H) independent experiments and two independent calculations (F). Data in I are pooled from two independent experiments, in which almost 150 cells at each time point were counted.