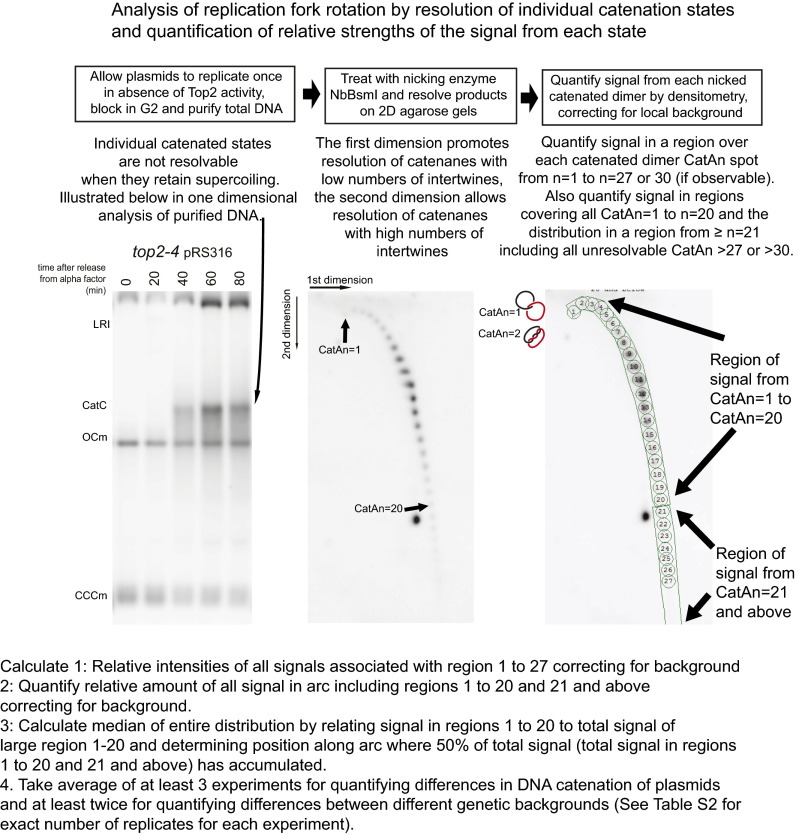
Fig. S2.
Quantification of fork rotation during DNA replication. In wild-type cells, Top2 resolves both precatenanes and catenanes formed during DNA replication. Therefore, to assay how often fork rotation and precatenation occur during replication on a plasmid replicon, we allowed one round of replication in the absence of Top2 activity and collected DNA from the cells blocked in G2 by nocodazole 80 min after release into the cell cycle (plasmid products are shown; Left). The catenated products of replication in this background are negatively supercoiled due to nucleosome deposition in the sister chromatids. This supercoiling of deproteinized plasmids normally compacts all catenated states into an unresolvable population. To resolve individual catenated states, the purified DNA is treated with a site-specific nicking enzyme to remove supercoiling but maintain the catenated nodes. The nicked DNA is resolved by agarose gel electrophoresis in two separate dimensions to be able to resolve both low- and high-catenated states before Southern blotting and probing for plasmid sequences (Middle). The relative intensities of each state were then calculated by densitometry in two ways. The first was by quantifying signal in equally sized regions centered on each catenated signal state on states 1–27 (or 1–30 for hypercatenated samples where the signal >27 was definable) and correcting for local background, and then expressing the relative intensity as a percentage of the total signal in all regions. The second measure was to compare the signal from the arc related to states 1–20 to states 21 and above. For this measure, regions were drawn around all states 1–20 and from the remainder of the arc relating to 21 and above. Then, each set of states is expressed as a percentage of the sum of both. This measure has the advantage of being able to quantify the signal from the individually unresolvable catenated states that produced detectable signal.