Fig. 1.
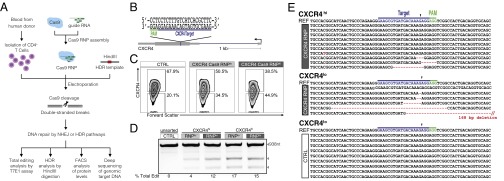
Efficient editing of CXCR4 in primary human CD4+ T cells. (A) Experimental scheme of Cas9:single-guide RNA ribonucleoprotein (Cas9 RNP) delivery to primary human CD4+ T cells for genome editing, followed by genetic and phenotypic characterization. (B) Schematic representation of sgRNA target (blue) and PAM (green) sequence designed to edit coding sequence in the human CXCR4 locus. (C) FACS plots show increasing percentages of cells with low CXCR4 expression (CXCR4lo) with higher concentrations of CXCR4 Cas9 RNP (Cas9 RNP lo: 0.9 µM; Cas9 RNP hi: 1.8 µM) compared with control-treated cells (Cas9 without sgRNA, CTRL; final concentration: 1.8 µM). (D) T7 endonuclease I (T7E1) assay demonstrates genome editing in the CXCR4 locus with more editing observed in FACS-sorted CXCR4lo cells than in CXCR4hi cells. Expected PCR product size (938 nt) and approximate expected sizes of T7E1-digested fragments are indicated. The total editing frequencies are indicated as percentage of Total Edit below the agarose gel image. (E) Mutation patterns detected by cloning and Sanger sequencing of the CXCR4 locus in sorted Cas9 RNP (1.8 µM)-treated CXCR4hi and CXCR4lo cells are compared with the sequence from CXCR4lo control-treated cells (CTRL). Reference (REF) sequence is shown on top of clonal sequences from each population with sgRNA target (blue) and PAM (green) sequences indicated. Red dashes denote deleted bases, and red sequences indicate mutated nucleotides. Arrowhead indicates the predicted Cas9 cut site. Poor quality sequences obtained from three additional CXCR4lo clones were removed from the sequence alignment.
