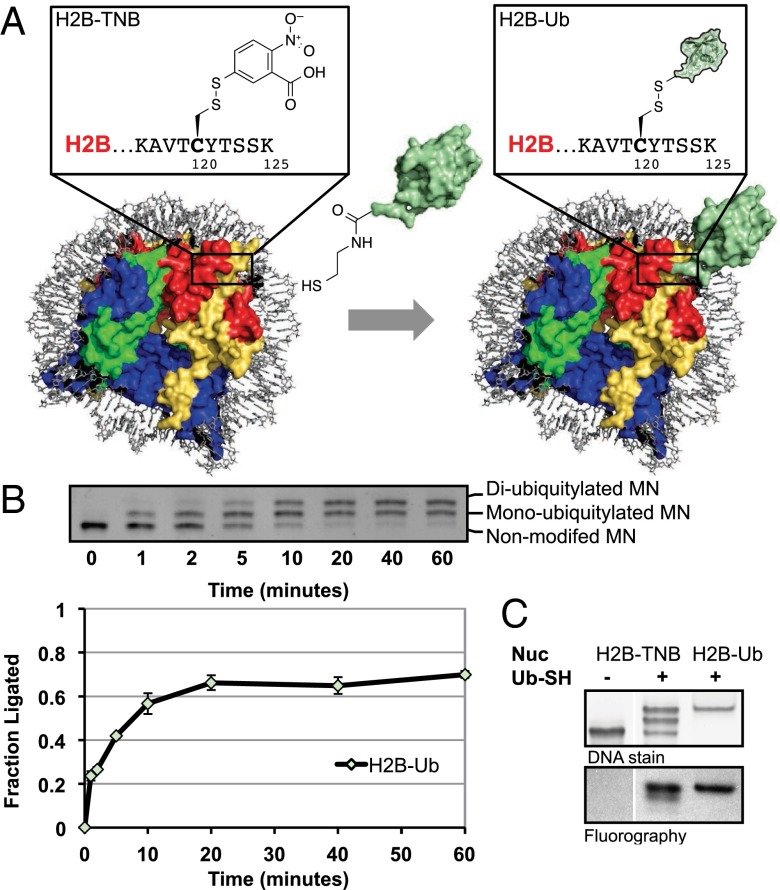
Fig. 1.
Preparation of H2B-Ub nucleosomes by on-nucleosomal asymmetric disulfide formation. (A) Scheme of nucleosomal ligation strategy. Site-specific ligation of ubiquitin (Ub) directly to the nucleosome is achieved upon addition of Ub-SH [unmodified ubiquitin shown in green; Protein Data Bank (PDB) ID code 1UBQ3] to H2B-TNB nucleosomes (wild-type nucleosome shown; PDB ID code 3LZ0). The H2B-Ub nucleosome is modeled as a composite of Ub and nucleosome structures. (Inset) Close-up of the H2B-TNB and H2B-Ub modifications. (B) Nucleosome ligations were monitored via native PAGE followed by ethidium bromide staining (Top panel) and quantified using ImageJ software (Bottom panel; see SI Appendix for quantification details). The reaction progressed to about 60% completion over the course of 1 h, resulting in a mix of non-, mono-, and diubiquitylated nucleosome species. Error bars, s.e.m. (n = 3). (C) hDot1L activity on H2B-Ub substrates prepared by nucleosomal ligations compared with fully reconstituted H2B-Ub substrates. hDot1L methyltransferase assays using 3H-SAM were performed on H2B-TNB nucleosomes (lane 1) and H2B-Ub nucleosomes, either prepared by direct nucleosomal ligations (lane 2) or fully reconstituted H2B-Ub (lane 3). Nucleosomes used in this assay were visualized via ethidium bromide staining of native PAGE gels (Top panel), and 3H-methyl incorporation was monitored by fluorography (Bottom panel). hDot1L was active on both types of H2B-Ub–containing nucleosomes but not H2B-TNB nucleosomes.