Figure 3.
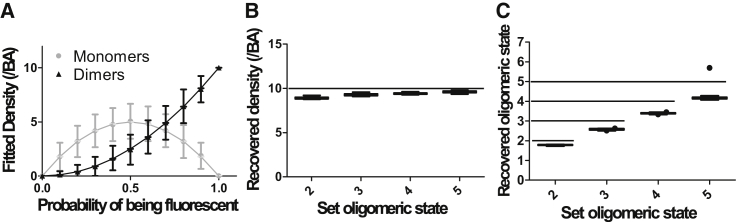
Effect of subunit mislabeling on SpIDA measurement of a uniform oligomeric state. (A) SpIDA results for simulated images containing only dimers as a function of labeling probability. Ten dimers where labeled with a subunit labeling probability p, and the distribution of observed fluorescent monomers and dimers was measured via SpIDA. This step was repeated 1000 times and the distribution of monomers and dimers as a function of the labeling probability is shown. The error bars correspond to SDs. The effect on the density (B) and quantal brightness (C) fits recovered with one-population SpIDA (Eq. 1) when only p = 80% of the oligomers are fluorescently labeled for different types of oligomers (nmer = 2–5). Here, we deliberately applied the wrong one-population SpIDA model without accounting for mislabeling (i.e., p = 100% is forced in the fit). The images generated for those simulations were 250 × 250 pixels, the oligomer density was set to 10 oligomers/BA, and p = 80%. (Box and whiskers) Tukey method; (dot) outliers.
