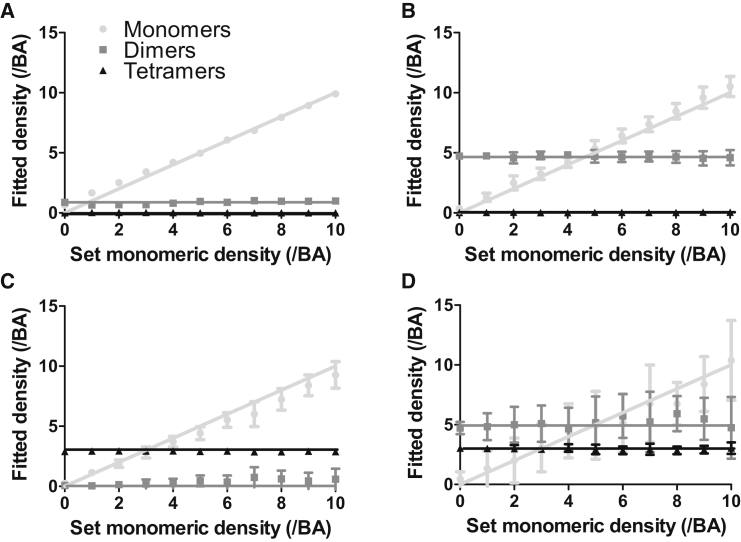
Figure 6.
Three-population SpIDA analysis with subunit mislabeling (p = 80%). SpIDA analysis was applied to simulated images containing three distinct populations (monomers, dimers, and tetramers) with 80% labeling of subunits. Here, the SpIDA results for many different cases where we set the density of dimers (N2) and tetramers (N4) and varied the number of monomers (N1) in the image from 0 to 10 per BA are shown. The results were obtained using Eq. 4 including mislabeling described in Eq. 5. (A) N2 = 1 dimer/BA, N4 = 0 tetramer/BA; (B) N2 = 5 dimers/BA, N4 = 0 tetramer/BA; (C) N2 = 0 dimer/BA, N4 = 3 tetramers/BA; and (D) N2 = 5 dimers/BA, N4 = 3 tetramers/BA. All the data points in the graphs correspond to averages of results from 20 images. The image size was 500 × 500 pixels. The error bars correspond to SDs.