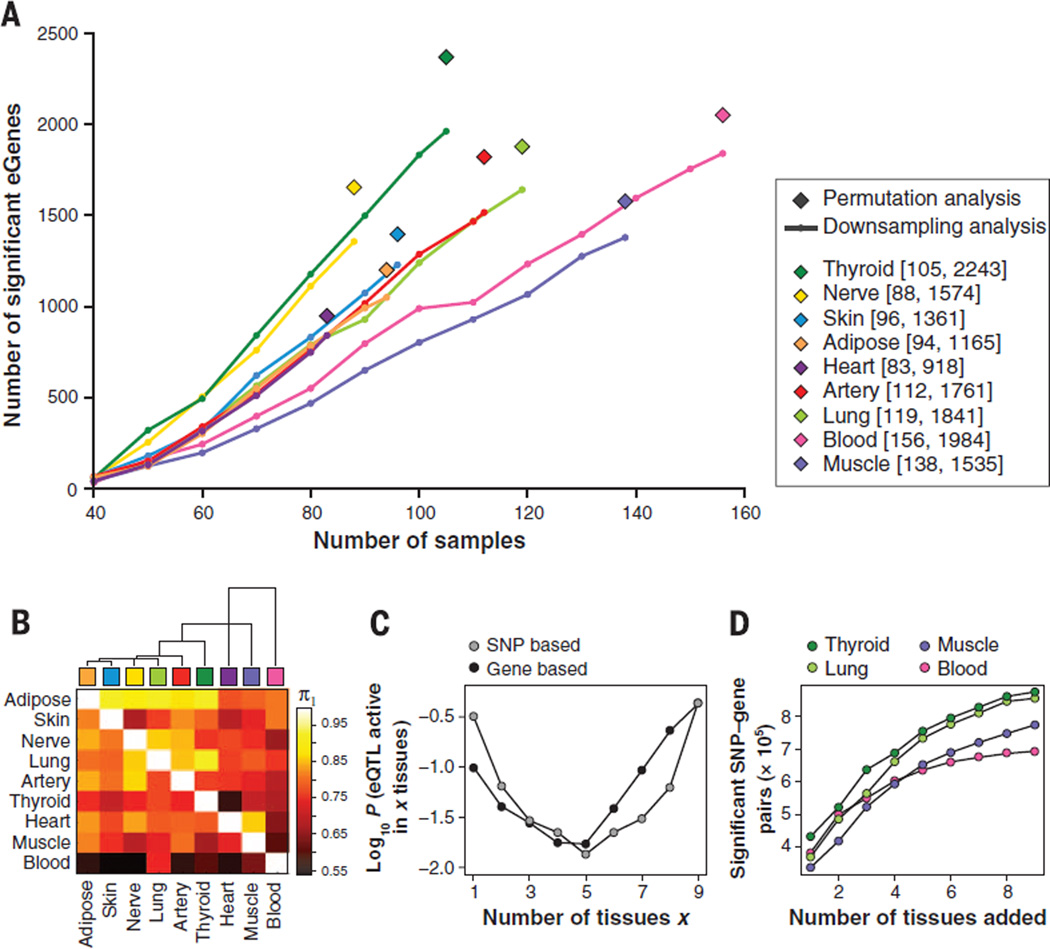
Fig. 2. Number and sharing of significant ciseQTLs per tissue.
(A) Numbers of significant cis-eQTL genes (eGenes) per tissue according to single-tissue analysis. For each gene, the minimum nominal P value was used as the test statistic and an empirical P value was computed to correct for number of tests per gene, based on either permutation analysis of genotype sample labels applied to the full set of samples per tissue (◆) or Bonferroni correction, used for downsampling (line) to reduce computational burden (14). In the range of sample sizes tested, the number of identified eGenes increases linearly with sample size. (B) Dendrogram and heat map of pairwise eQTL sharing using the method of Nica et al. (22). Values are not symmetrical, since each entry in row i and column j is an estimate of π1 = Pr(eQTL in tissue i given an eQTL in tissue j). Blood has the lowest levels of eQTL sharing with other tissues while adipose shows higher levels of sharing. (C) Activity probabilities for both multitissue modeling approaches, applied to all nine tissues, indicate that the most likely configurations are for eQTLs that are active in only a few tissues or in many tissues. (D) For eQTLs in each tissue considered separately, analyzing multiple tissues jointly increases the number of discovered eQTL associations (FDR < 0.05), as assessed by the SNP-based multitissue model.