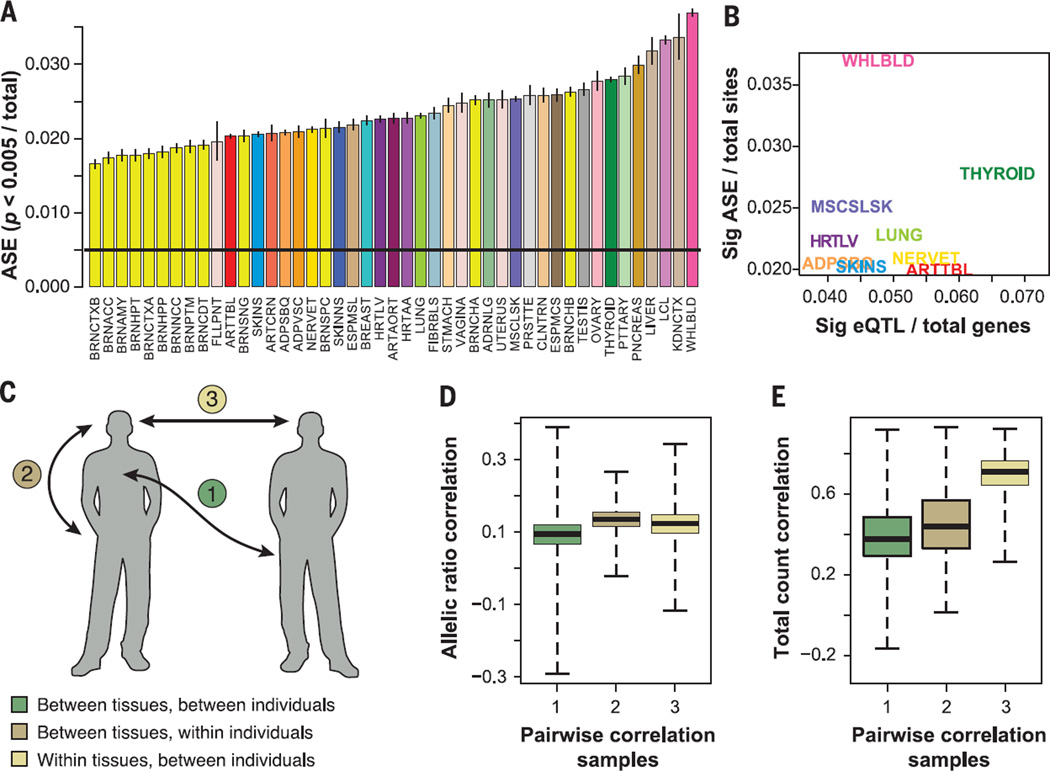
Fig. 3. Quantification of regulatory diversity by ASE.
(A) Proportion of sites with significant ASE (P < 0.005) in each tissue (colored and labeled as in Table 1), with binomial confidence intervals. (B) Proportion of significant ASE sites for the nine tissues with eQTL data as a function of the proportion of eQTLs after regressing out the log of sample size. (C) Partitioning variation in allelic and total gene expression within and between individuals and tissues. We calculated pairwise Spearman rank correlations between all the samples using two metrics [(D) and (E)]. (D) Allelic ratios over sites (sampled to 30 reads each), which captures similarity in allelic effects that are a proxy for cis-regulatory variation. (E) Total read counts over the same sites, which captures similarity in total gene expression levels. The plots show the distributions of pairwise correlations for sample pairs that are from (1) different tissues and different individual, (2) different tissues within an individual, or (3) same tissues in different individuals. Gene expression levels are highly correlated within the same tissue (E3) (see Fig. 1A). However, allelic ratios show highest correlation among different tissues of the same individual that share the same genome (D2).