Fig. 1.
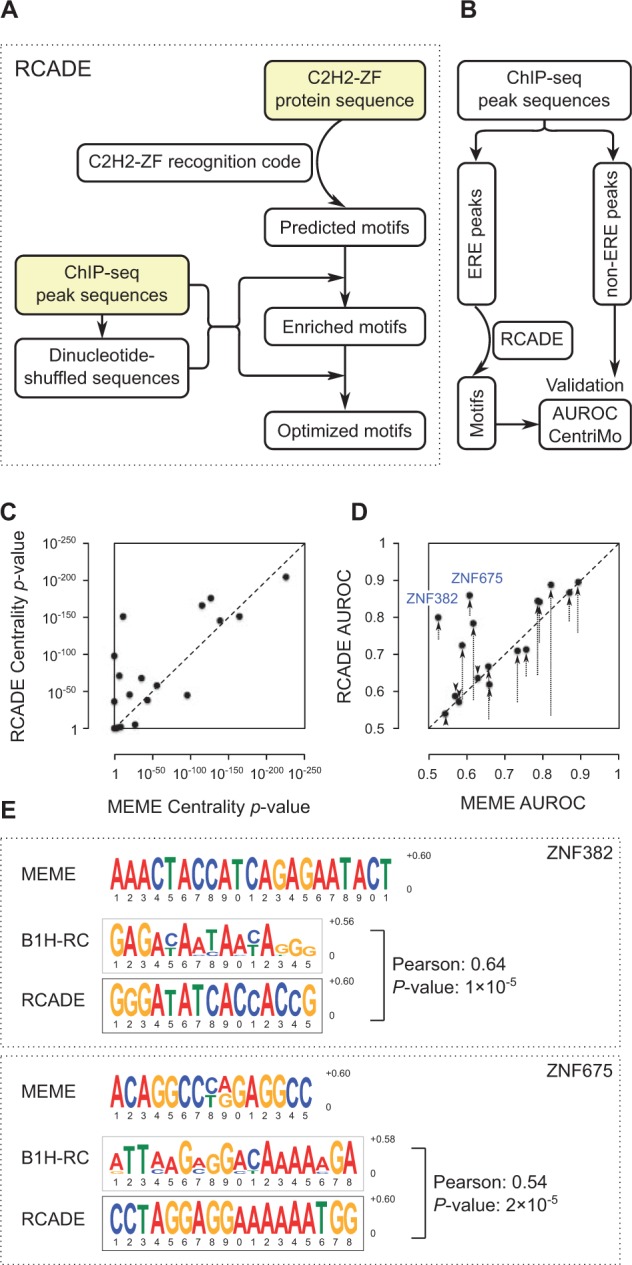
RCADE workflow and benchmarking results. (A) RCADE starts by predicting a set of motifs from the target C2H2-ZF protein sequence, using a previously published bacterial-one-hybrid assay-based recognition code, or B1H-RC (Najafabadi et al., 2015), which are evaluated against the ChIP-seq peak sequences to identify significantly enriched motifs, and are then iteratively optimized. (B) Benchmarking workflow for evaluation of RCADE. The peak sequences were divided into two sets of ERE-overlapping and non-ERE peaks. The ERE-overlapping peaks for each protein were used for motif discovery using RCADE, and the motifs were validated using non-ERE peaks. (C,D) Validation results for 18 ERE-binding proteins. The arrows show the improvement in the AUROC of RCADE motifs compared with seed B1H-RC motifs. (E) Example motifs for two proteins that show the largest difference between RCADE and MEME validation results. The top-scoring MEME motif is shown for each protein, followed by the top-scoring motif that is directly predicted from protein sequence using the B1H-RC, and the RCADE optimized motif. The Pearson similarity of the B1H-RC and RCADE motifs was calculated as described previously (Najafabadi et al., 2015)
