Figure 3.
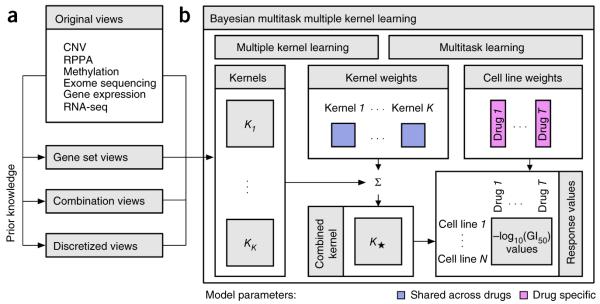
The method implemented by the best performing team. (a) In addition to the six profiling data sets, three different categories of data views were compiled using prior biological knowledge, yielding in total 22 genomic views of each cell line. (b) Bayesian multitask MKL combines nonlinear regression, multiview learning, multitask learning and Bayesian inference. Nonlinear regression: response values were computed not directly from the input features but from kernels, which define similarity measures between cell lines. Each of the K data views was converted into an N×N kernel matrix Kk (k = 1,…,K), where N is the number of training cell lines. Specifically, the Gaussian kernel was used for real-valued data, and the Jaccard similarity coefficient for binary-valued data. Multiview learning: a combined kernel matrix K* was constructed as a weighted sum of the view-specific kernel matrices Kk, k = 1,…,K. The kernel weights were obtained by multiple kernel learning. Multitask learning: training was performed for all drugs simultaneously, sharing the kernel weights across drugs but allowing for drug-specific regression parameters, which for each drug consisted of a weight vector for the training cell lines and an intercept term. Bayesian inference: the model parameters were assumed to be random variables that follow specific probability distributions. Instead of learning point estimates for model parameters, the parameters of these distributions were learned using a variational approximation scheme.
