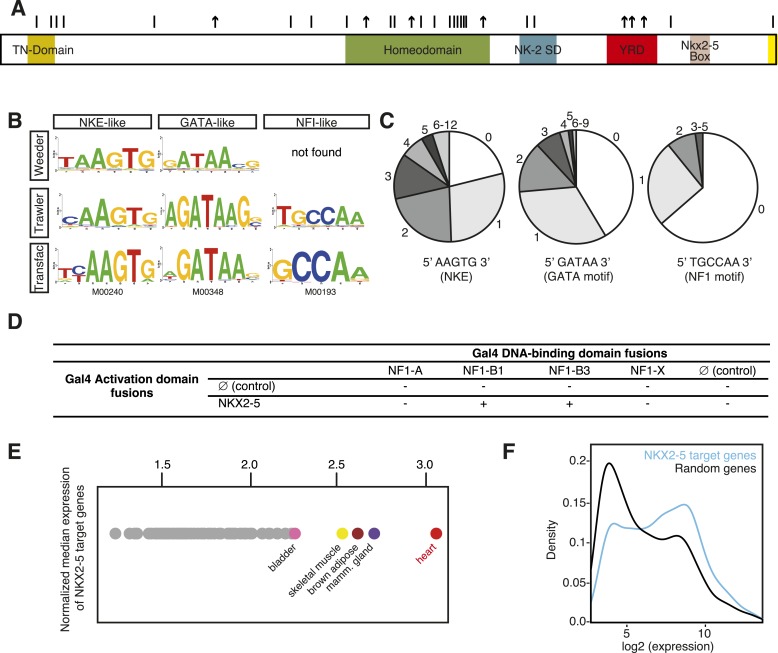
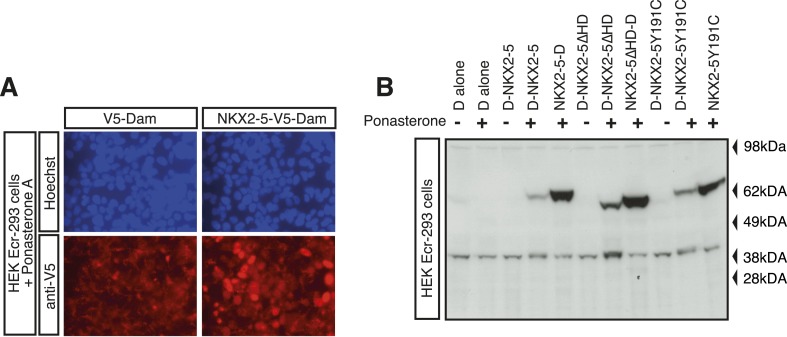
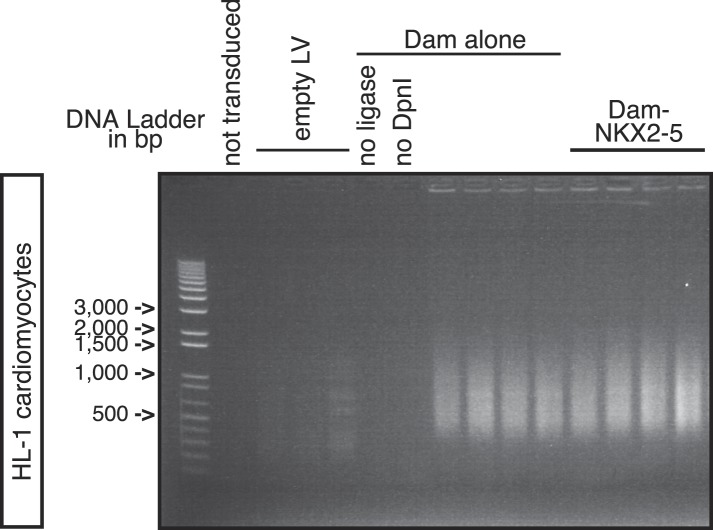
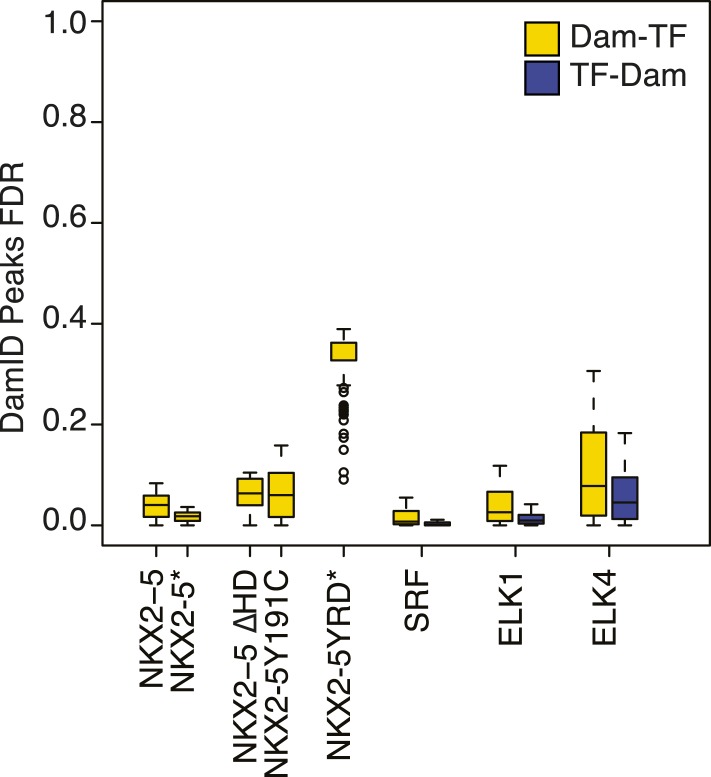
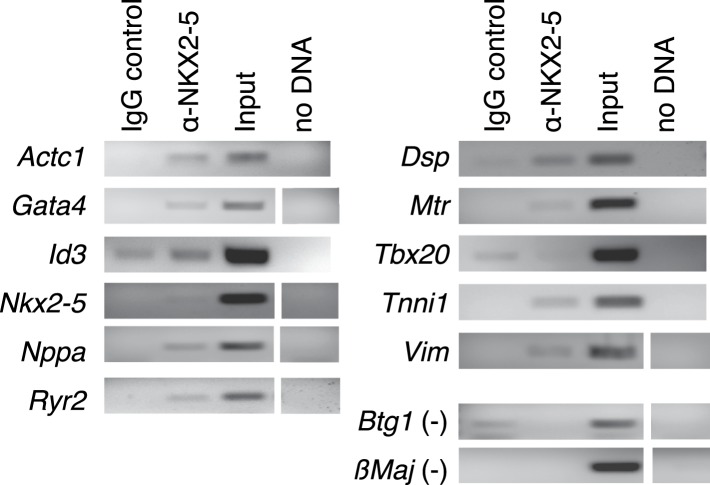
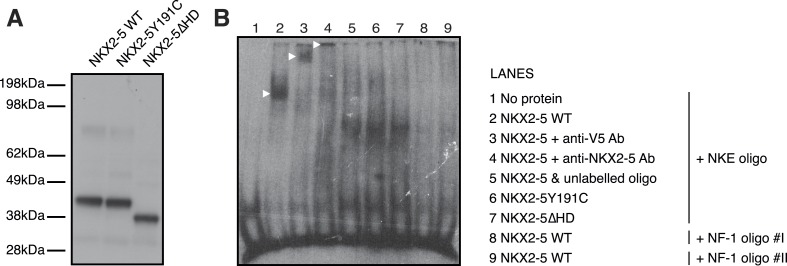
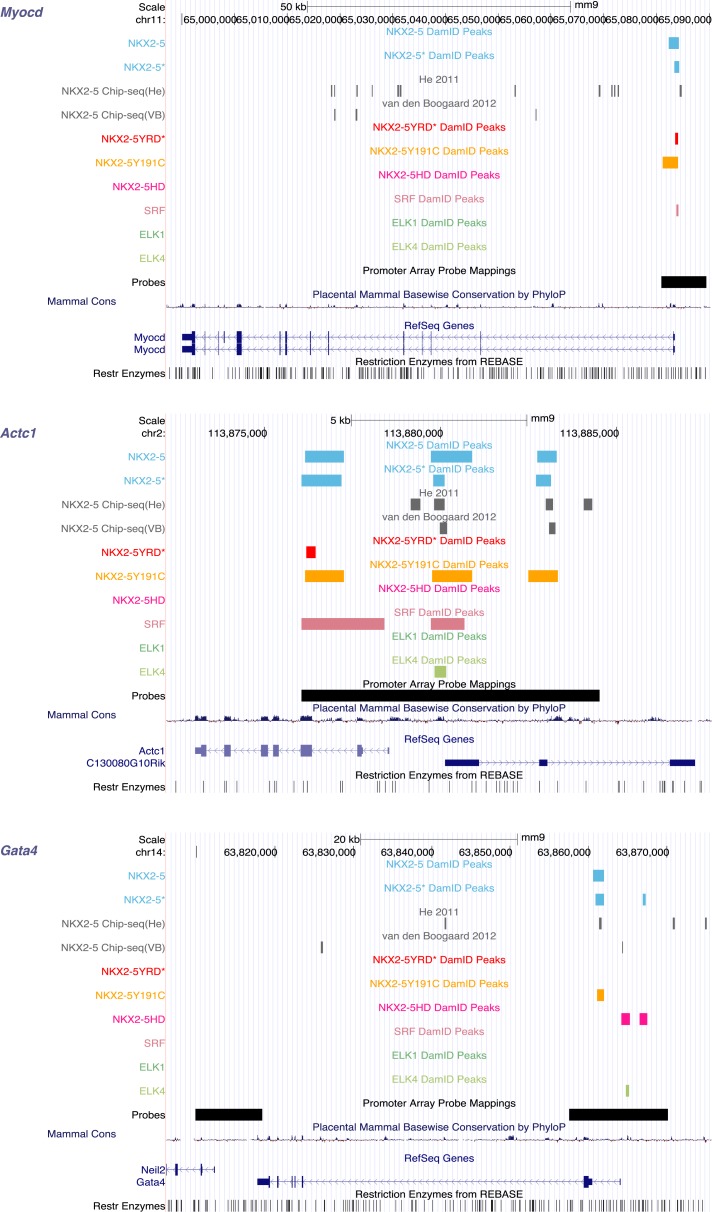
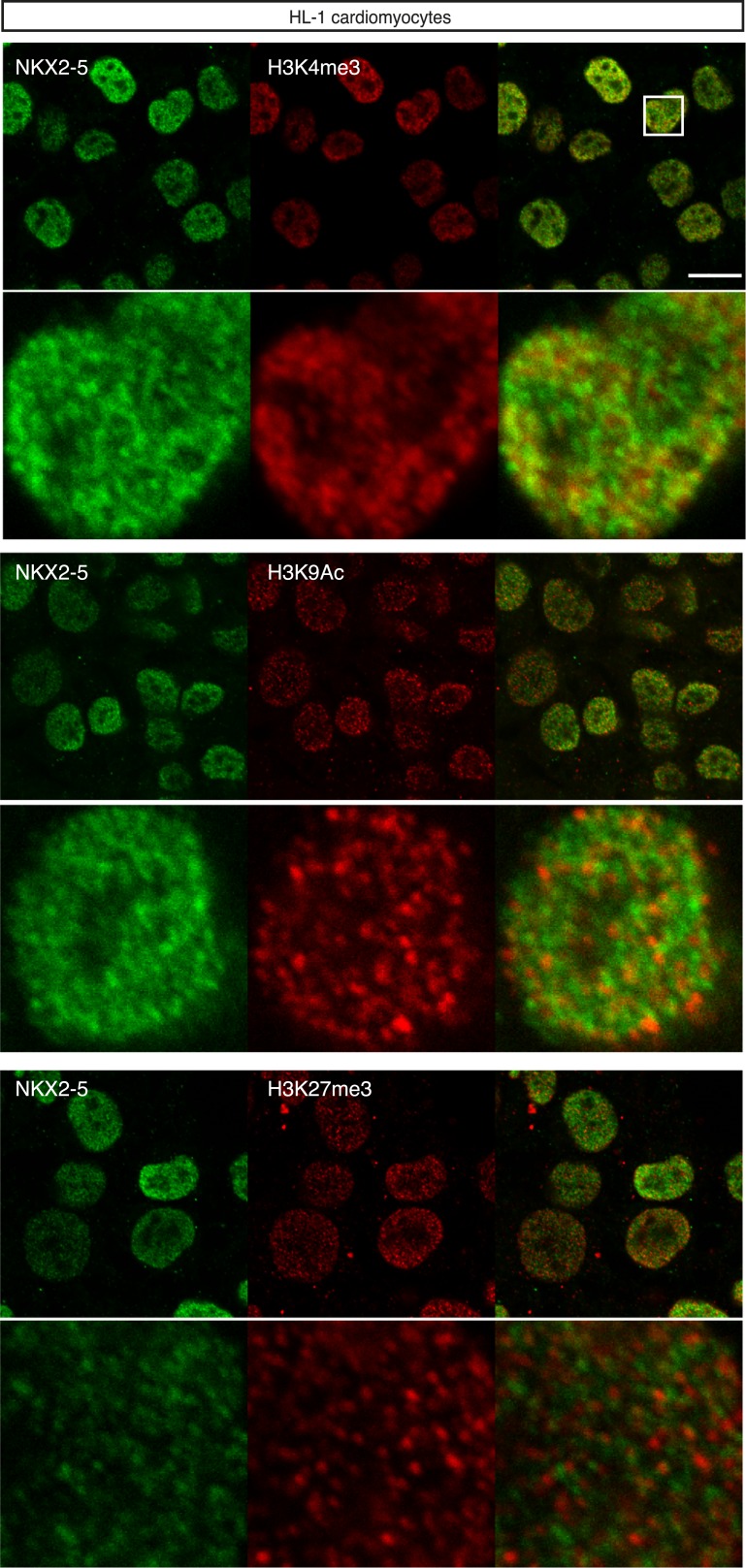
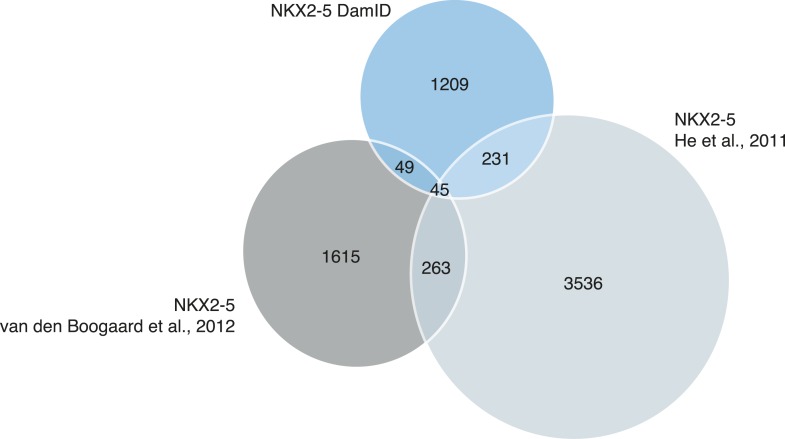
Figure 1. DNA adenine methyltransferase identification (DamID) identifies a robust set of NKX2-5 targets in HL-1 cardiomyocytes.
(A) Structure of the human NKX2-5 protein (TN, tinman domain; NK2SD, NK-2 specific domain; YRD, tyrosine-rich domain). Bars and arrows indicate missense and termination mutations associated with congenital heart disease (CHD), respectively. (B) Top over-represented motifs discovered de novo in NKX2-5 peaks using Trawler or Weeder. NKX2-5, GATA, and Nuclear Factor 1 (NF1) binding motifs deposited in TRANSFAC are shown. (C) Distribution of NKX2-5, GATA, and NF1 binding sequences in NKX2-5 peaks. (D) Yeast-two-hybrid assay. NKX2-5 and NF1 proteins were fused to Gal4-activation and DNA-binding domains, respectively. Positive signs (+) show interaction as growth on selective medium from three independent experiments. (E) Normalized median expression of NKX2-5-target genes in 91 murine cell types (data collected from BioGPS). Tissues with the highest median expressions are shown in colour, including heart (red). (F) Expression of NKX2-5 target genes and random genes in HL-1 cells. Data collected from (Mace et al., 2009).