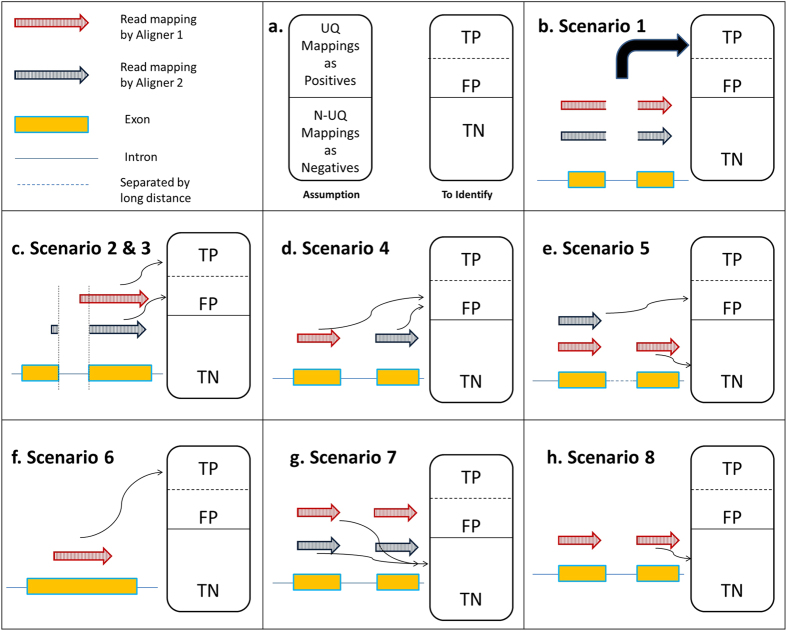
Figure 1. The core algorithm of CADBURE.
(a) The assumption of CADBURE core algorithm and metrics to identify. The collection of UQ (unique) mappings are assumed as a set of positives and the collection of N-UQ (non-unique) mappings are assumed as a set of negatives. (b) Scenario 1. UQ mapping by both aligner-1 (red arrow) and aligner-2 (blue arrow) to the same genome location with the same alignment. (c) Scenario 2 & 3. UQ mapping by both aligner-1 and aligner-2 to overlapping genome regions with different alignments. Scenario 2: No spliced alignment by aligner-1 is called as TP (true-positive). Scenario 3: spliced-alignment by aligner-2 with a nucleotide overhang is called as FP (false-positive). (d) Scenario 4. UQ mapping by both aligner-1 and aligner-2 to different genomic locations, and both are called FP. (e) Scenario 5. UQ mapping by aligner-2 is called FP because the same read is reported as N-UQ mapping by aligner-1. The read mapping in the aligner-1 is called TN (true-negative). (f) Scenario 6. UQ mapping by aligner-1 is called TP because aligner-2 cannot map this read as either UQ or N-UQ mapping. (g) Scenario 7. N-UQ mapping by both aligner-1 and aligner-2 where they agree for a read in that they map it non-uniquely. This case is called TN for both aligners. (h) Scenario 8. N-UQ mapping by aligner-1 is called TN because aligner-2 cannot map this read as either UQ or N-UQ read. UQ: Unique, N-UQ: Non-Unique, TP: true-positive, FP: false-positive, TN: true-negative.