Fig. 4.
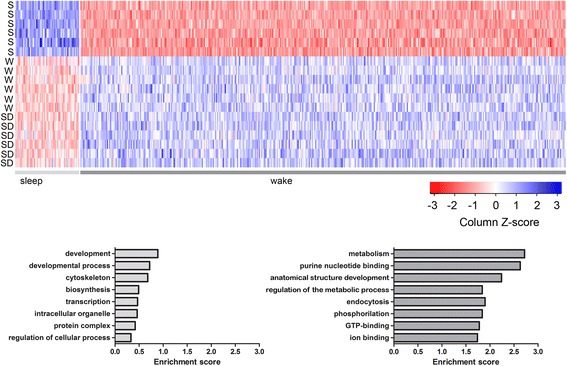
Functional characterization of genes differentially expressed in sleep (S) and wake (W + SD). Top: Heat diagrams show the probeset intensity for each individual animal in the three experimental conditions. Bottom: Functional annotation analysis (DAVID default settings, except for kappa = 4, similarly threshold = 0.7) for S (n = 55) and W + SD (n = 396) genes. The top eight functional annotation clusters in order of enrichment score are shown for S (left) and W + SD (right)
