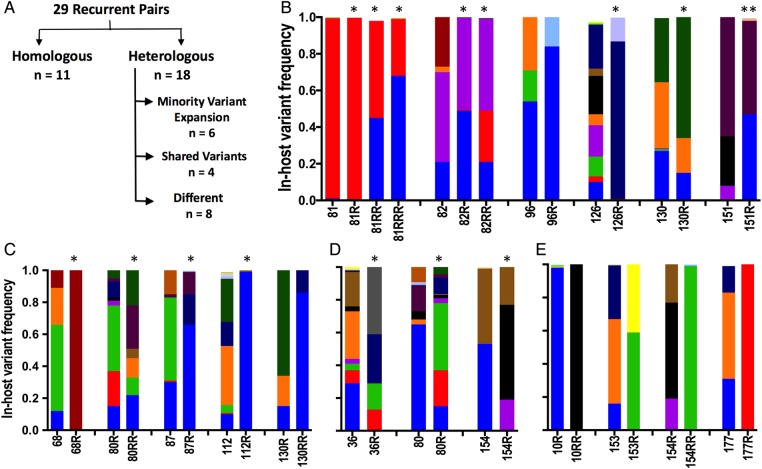
Figure 3.
Representative genotypes of recurrent pairs categorized into homologous or heterologous pairs (A) based on overlap of pvmsp1 variants in the recurrent and preceding infection. Unique haplotypes are represented by specific colors across all samples. B, Homologous pairs were defined as having the same dominant or codominant haplotype at recurrence as seen in the preceding episode. C, In pairs exhibiting minority variant expansion, a minority parasite population existing at <20% in-host frequency in the initial infection reappeared as the dominant variant at recurrence. D, At least 1 shared variant defined our third category, while one-third of heterologous pairs shared no overlap at all (E). Pairs identified as probable relapses based on statistical testing are denoted by an asterisk. A pair that was “indeterminate” by statistical testing but judged as likely relapse based on microsatellite results are denoted by double asterisks. The genotypes of all 29 pairs are depicted in Supplementary Figure 3 (21 depicted here).