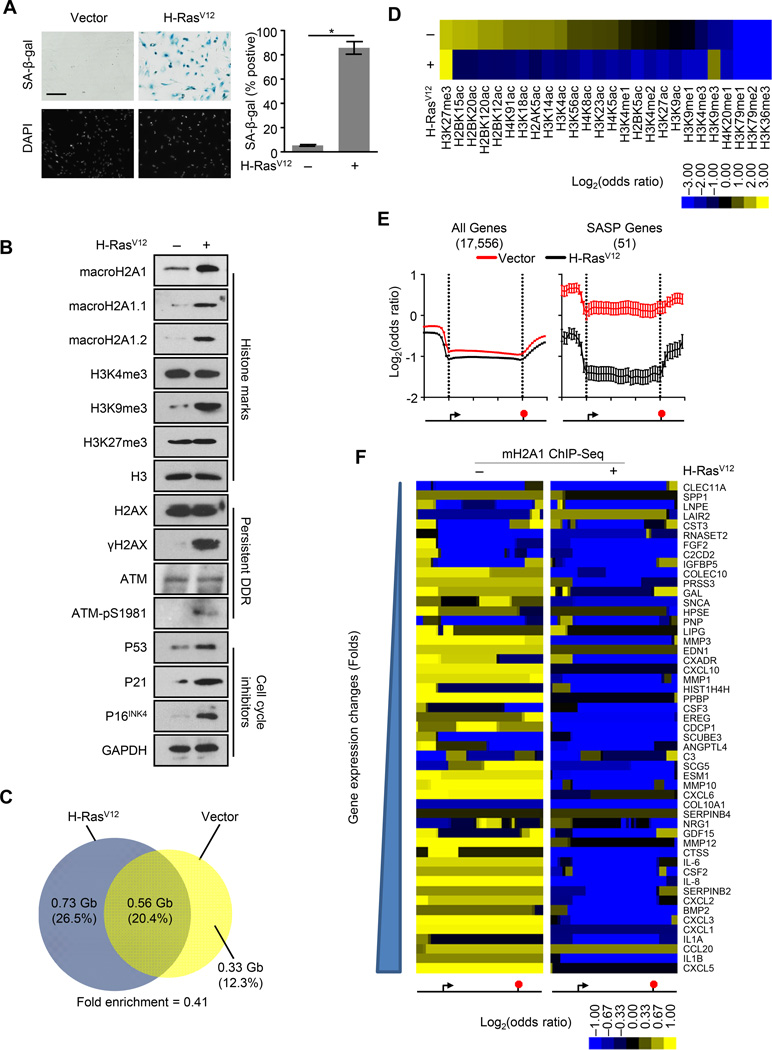
Figure 1. MacroH2A1 undergoes large-scale genome-wide rearrangement upon OIS.
(A) Left, representative SA-β-gal and DAPI counterstaining of IMR90 cells subjected to retroviral-mediated expression of H-RasV12 or empty vector as a control for 14 days. Scale bar represents 200 µm. Right, histogram depicting the percentage of SA-β-gal positive cells. Error bars, s.e.m. (n = 3 independent cell passages and retroviral infections). *p < 0.05 from two-tailed Student’s t-tests.
(B) Immunoblots for macroH2A1, various histone marks, DDR factors and cell cycle inhibitors from IMR90 cells treated as in (A).
(C) Two-way Venn diagrams depicting the overlap between ChIP-seq identified bound regions for macroH2A1 in IMR90 cells treated as in (A). Numbers indicate the DNA coverage (Gb) and percentage of DNA in each section.
(D) Heat map depicting the log2(odds ratio) for the overlap between macroH2A1-enriched domains identified in IMR90 cells treated as in (A) and 26 histone PTMs.
(E) Metagene analysis of ChIP-seq data for macroH2A1 from IMR90 cells treated as in (A). Left, all autosomal protein coding genes. Right, fifty-one SASP genes defined from gene expression microarray data. The data represent the average signal in ten 1-kb windows upstream of the TSS, 30 windows spanning the gene body and ten 1-kb windows downstream of the end of the gene. The location of the TSS and the end of the gene were depicted by the left and right vertical dotted line respectively. Error bars, s.e.m. (n = 17,556 and 51 for red and black curves respectively).
(F) Heat map of macroH2A1 occupancy in IMR90 cells treated as in (A) for the 51 individual SASP genes. The data are represented as ten 1 kb windows upstream of the TSS, 30 windows spanning the gene body, and ten 1 kb windows downstream of the end of the gene. See also Figure S1.