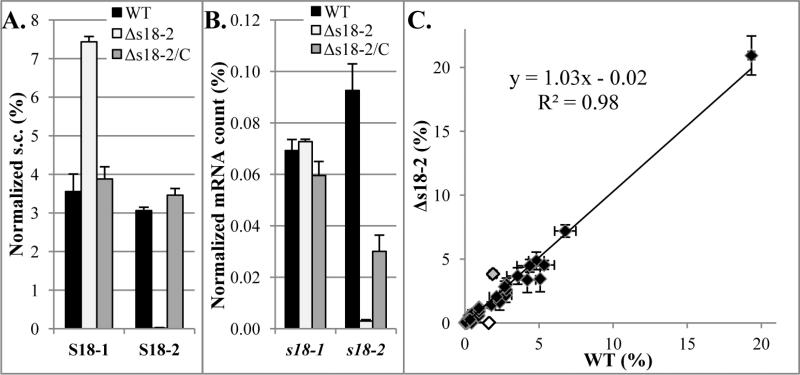
FIGURE 5. Changes in protein and mRNA levels of S18-1 in the presence or absence of S18-2.
Wild type (WT), deletion mutant (Δs18-2), and complemented (Δs18-2/C) strains of M. tuberculosis H37Rv were grown to stationary phase in Sauton's medium and harvested for protein and RNA quantification.
A. Protein: Mass spectrometry analysis Spectral count (s.c.) for identified peptides normalized against s.c. of all identified ribosomal proteins
B. mRNA: qRT-PCR of S18 genes mRNA normalized against 16S rRNA. Note that the complemented strain does not have wild type levels of s18-2 mRNA, but complements well at the protein level.
C. Comparison of proteins in wild type (WT) and deletion mutant (Δs18-2) strains
Spectral counts of identified proteins were normalized to total spectral counts in each sample. Gray data point is the S18-1 protein and white is the S18-2 protein. The other three AltRPs from the same operon were also detected and their amount was not changed in the absence of the S18-2 protein. Note that only the 10-15kDa range was analyzed. Error bars in all graphs are ±1 standard deviation of technical duplicates.