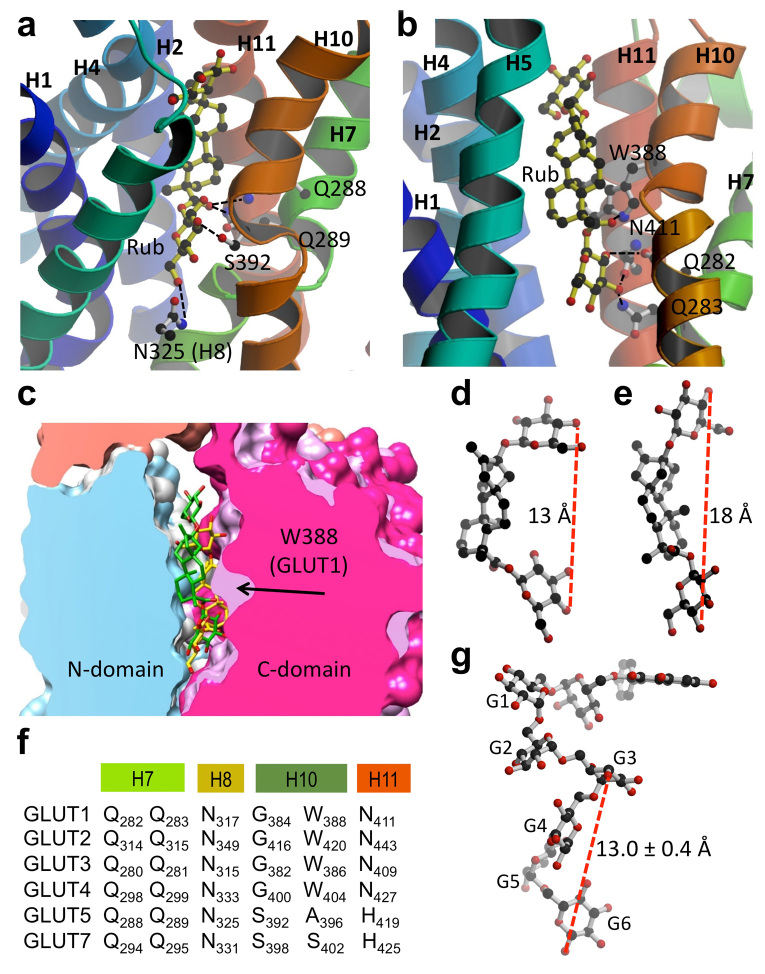
Figure 3. Rub binding to GLUTs.
(a) and (b) Cytosolic face of GLUT is oriented towards the top, and helices 8 and 9 have been removed for better visualization. Images were generated with MolScript32 and raster3D33. (a) Modeled GLUT5 with docked Rub. Predicted Rub-interacting residues are: Q288 and Q289 from helix 7, N325 from helix 8, and S392 from helix 10. (b) GLUT1 (PDB ID 4PYP) with docked Rub. Predicted interacting residues are: Q282 and Q283 from helix 7, W388 from helix 10, and N411 from helix 11. (c) Comparison of space-filling models of GLUT1 (white in N-domain, light purple in C-domain) and GLUT5 (blue in N-domain, magenta in C-domain) interacting with Rub (shown as stick molecule, in yellow for GLUT1, and green for GLUT5). Molecular graphics and analyses were performed with the UCSF Chimera package34. (d) Rub as it docked to GLUT5 binding site. The distance between 4-oxygen positions of the end glucosides is 13 Å. (e) Rub as it docked to GLUT1 binding site. The distance between 4-oxygen positions of the two glucosides is 18 Å. (f) Alignment of GLUT1-5, 7 residues predicted to interact with Rub. (g) Model of Ast6G, with the astragalin head modeled onto isomaltooctaose chain from PDB ID 3WNN.