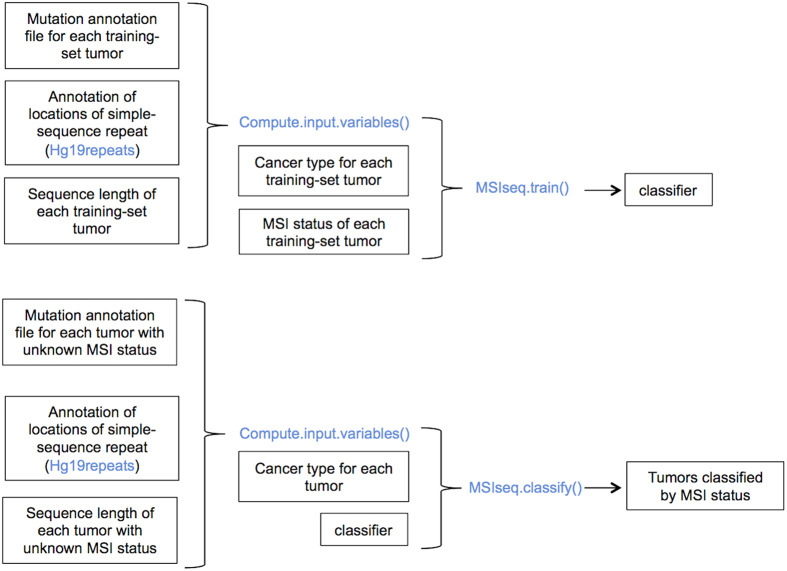
Figure 5. Workflow for the R MSIseq package.
Functions and variables in the package are highlighted in blue. MSIseq provides Compute.input.variables() to calculate the potential input variables (S.ind, T.sns, etc.) from (i) a mutation annotation file, (ii) an annotation of the locations of simple repeats in the genome, and (iii) the lengths of the sequenced regions of the genome that were searched for somatic mutations. MSIseq provides these data as used in this paper in the variables NGStraindata, Hg19repeats, and NGStrainseqLen. MSIseq.train() takes the input variables plus (optionally) cancer type information and creates a classifier. Please refer to the MSISeq documentation and vignette for details. MSIseq also provides a pre-computed classifier (called NGSclassifier in the package) that implements the NGSclassifier presented in this paper. For classification of samples with unknown MSI status, input variables can be prepared from the mutation annotation file by Compute.input.variables() and then passed to MSIseq.classify() along with a classifier generated by MSIseq.train().