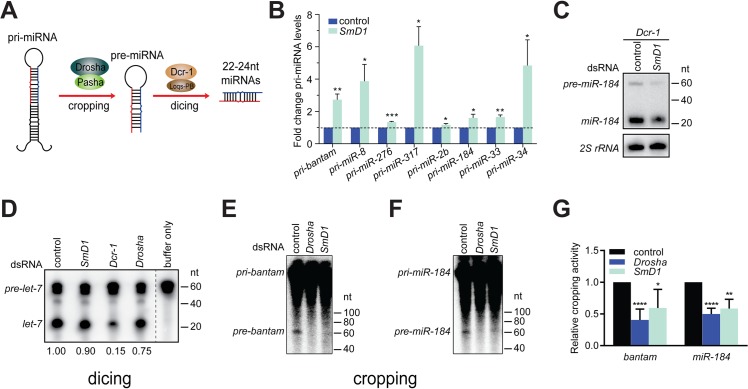
Fig 2. Depletion of SmD1 compromises microprocessor activity.
(A) A schematic of the cropping and dicing steps of miRNA biogenesis. (B) Levels of various primary miRNA transcripts in SmD1-depleted cells or control cells were measured by RT-qPCR and normalized against the control rp49 mRNA (n ≥ 3). (C) S2 cells were treated with control or SmD1 dsRNAs together with the Dcr-1 dsRNA, and levels of the precursor and mature miR-184 were measured by Northern blot. (D) Cytoplasmic lysates from S2 cells treated various dsRNAs were incubated with radiolabeled pre-let-7 substrate to generate mature let-7. RNAs were extracted and resolved by denaturing urea-PAGE. Quantification of Dcr-1 activity is shown at the bottom. (E,F) Total lysates from S2 cells treated various dsRNAs were incubated with radiolabeled pri-bantam (E) or pri-miR-184 (F) to generate pre-miRNAs. (G) The microprocessor activity, represented as the amount of resultant pre-miRNAs, was quantified and normalized to controls (n ≥ 4).