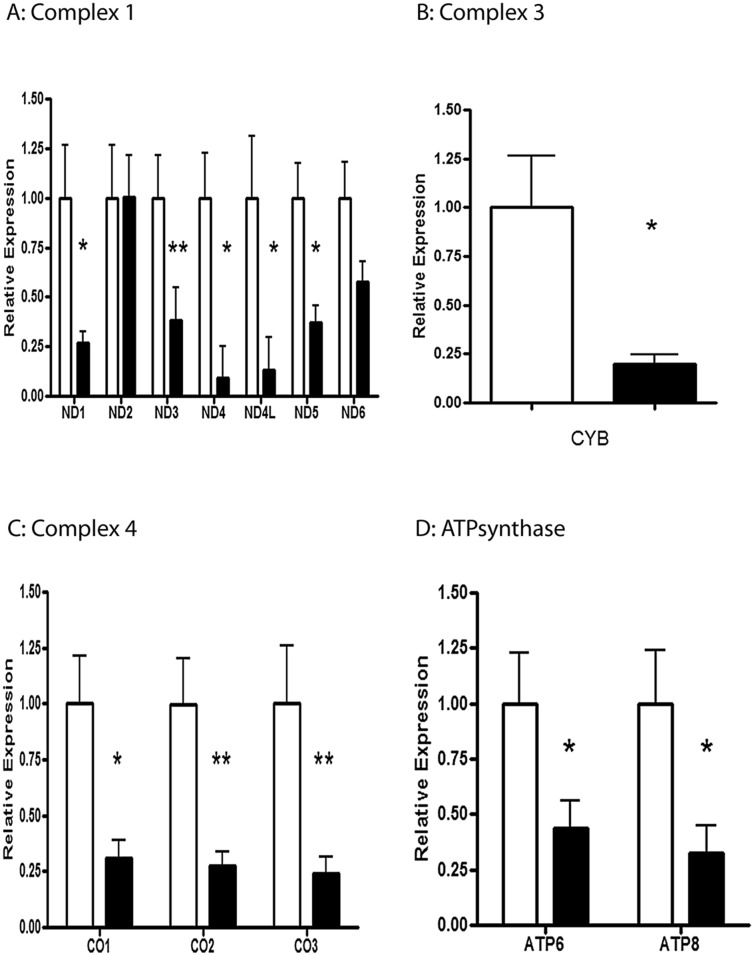
Fig 2. Quantitative Real-Time PCR mt-genes across 4 complexes of ETC.
A: mt-ND1 was reduced ~3.7 fold in HT BN/SHR-mtSHR (*P<0.05). mt-ND3 was reduced ~2.6 fold in HT SHR/BN-mtSHR (**P<0.01). mt-ND4 was reduced ~10.8 fold in HT SHR/BN-mtSHR (*P<0.05). mt-ND4L was reduced ~7.7 fold in HT SHR/BN-mtSHR (P<0.05). mt-ND5 was reduced ~2.7 fold in HT SHR/BN-mtSHR (P<0.05). mt-ND6 was reduced ~1.7 fold in HT SHR/BN-mtSHR (P>0.05) (Fig 1A). mt-ND2 was not different between the two phenotypes (P>0.05). B: mt-CYB was reduced ~5 fold in HT BN/SHR-mtSHR (*P<0.05). C: mt-CO1 was reduced ~3.2 fold in HT BN/SHR-mtSHR (*P<0.05), mt-CO2 was reduced ~3.6 fold in HT BN/SHR-mtSHR (P<0.05), and mt-CO3 was reduced 4.1 fold in HT BN/SHR-mtSHR (P<0.05). D: mt-ATP6 was reduced ~2.3 fold in HT BN/SHR—mtSHR (*P<0.05), while mt-ATP8 was reduced ~3.1 fold in HT BN/SHR-mtSHR (*P<0.05). (NT: open bars; HT: closed bars).