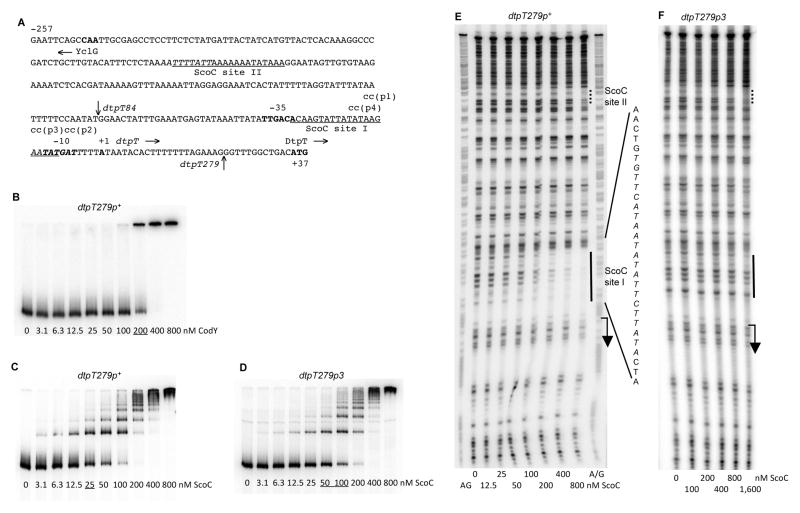
Fig. 3.
Binding of CodY and ScoC to the dtpT regulatory region.
A. The sequence (5’ to 3’) of the coding (non-template) strand of the dtpT regulatory region. Coordinates are reported with respect to the transcription start point as deduced from the mutational analysis (see text). The 5’ nucleotide of the sequence presented corresponds to the first nucleotide of the dtpT insert within the dtpT279-lacZ fusions. The vertical arrow above the sequence indicates the 5’ nucleotide of the dtpT84-lacZ fusion at position −62. The vertical arrow below the sequence indicates the junction point, at position +22, between the dtpT and lacZ sequences. The likely translation initiation codon (ATG), −10 and −35 promoter regions, and transcription start point are in bold. The directions of transcription and translation are indicated by the horizontal arrows. The mutated nucleotides are shown in lowercase above the sequence. The sequences protected by ScoC in DNase I footprinting experiments on the template strand of DNA are underlined. The ScoC-binding motifs are italicized.
B, C and D. Gel shift assay of protein binding to a radioactively labeled dtpT279p+ (B and C) or dtpT279p3 (D) PCR fragment, obtained with oligonucleotides oBB67 and oBB102. The proteins tested for binding were (B) CodY in the presence of 10 mM ILV or (C and D) ScoC. Protein concentrations (nM of monomer) are reported below each lane. The underlined concentrations indicate the apparent KD for binding.
E and F. DNase I footprinting analysis of ScoC binding to the dtpT regulatory region. The dtpT279p+ (E) or dtpT279p3 (F) DNA fragment labeled on the template strand and used in panels B or D, respectively, was incubated with increasing amounts of purified ScoC and then treated with DNase I. See the legend to Fig. 1E for additional details.