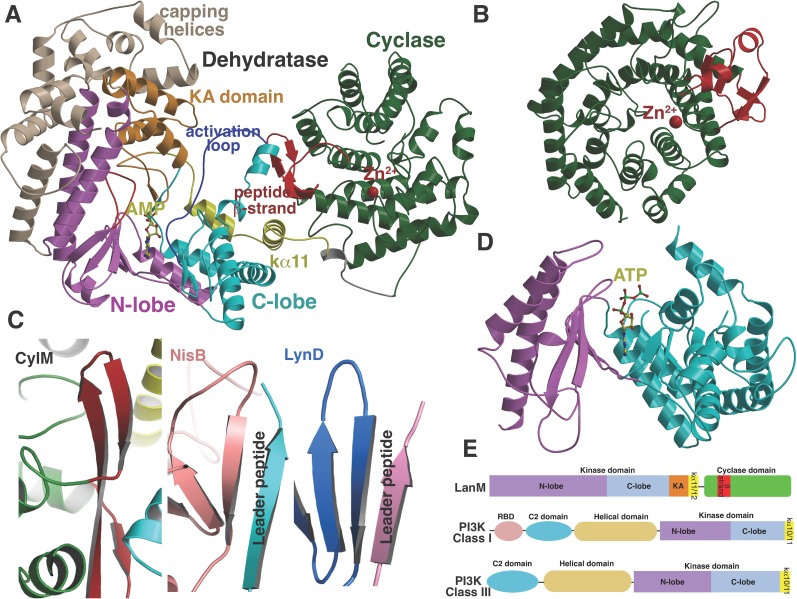
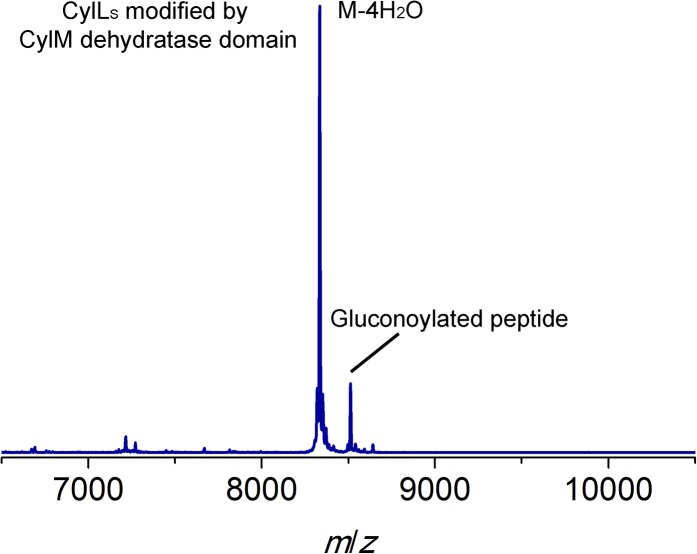
Figure 2. (A) Overall structure of CylM.
(B) Structure of the class I lanthipeptide cyclase NisC illustrating the structural homology with the C-terminus of CylM. (C) Comparison of the putative peptide-binding β-strands of CylM with the peptide binding regions of other RiPP biosynthetic enzymes including NisB (involved in nisin biosynthesis, PDB 4WD9) and LynD (involved in cyanobactin biosynthesis; PDB 4V1T). (D) Structure of the lipid kinase PI3K that shares homology with the dehydration domain of CylM. (E) Domain organization of LanMs in comparison with that of lipid kinases. RBD, Ras-binding domain.