Figure 2.
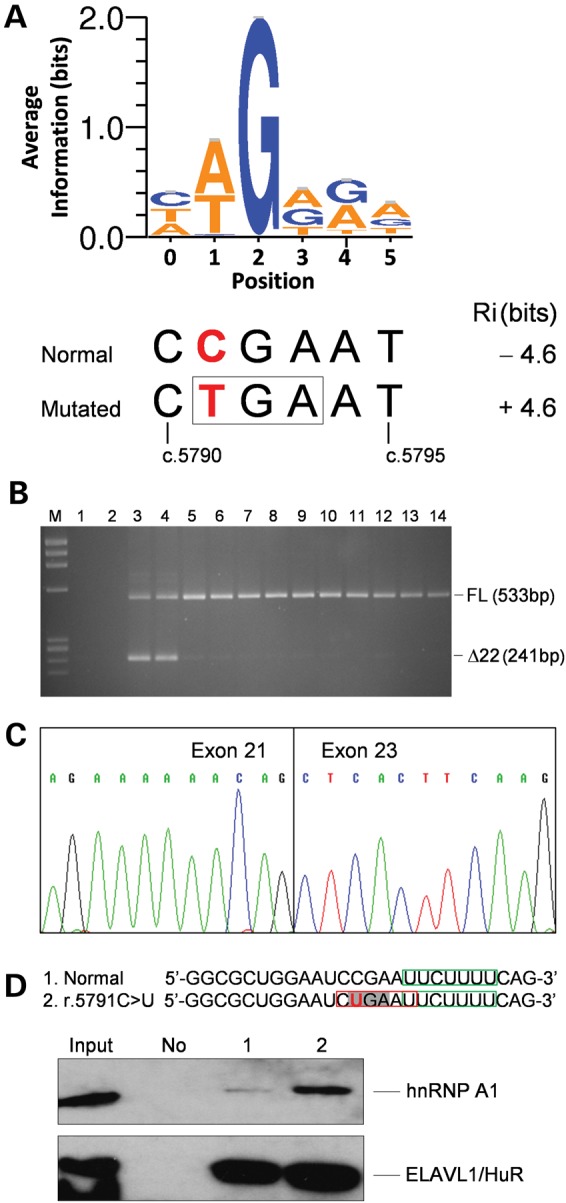
Analysis of the effect of the FANCM c.5791C>T mutation on RNA. (A) Sequence logo of hnRNP A1 binding sites generated as described in Materials and Methods. The opal codon (TGA, boxed) introduced by the FANCM c.5791C>T mutation (in bold red) is contained at positions 1–3 of the hnRNP A1 binding-site encompassing nucleotides c.5790_5795. The hnRNP A1 binding-site strength computed by the ASSEDA software for the normal and mutated sequences is reported. (B) Agarose gel electrophoresis of the RT-PCR products using a forward primer in exon 21 and a reverse primer in exon 23. M, molecular marker (ΦX-174 HaeIII digested); 1, no template as a negative control for PCR; 2, genomic DNA as a negative control for the specificity of cDNA amplification; 3 and 4, cDNAs from LCLs carrying the c.5791C>T mutation; 5–14, cDNAs from LCLs derived from 10 mutation negative individuals, used as reference controls. The sizes of the full-length (FL) and Δexon22 (Δ22) transcripts are indicated. (C) Sequence of the aberrant band excised from the gel showing the skipping of the entire exon 22. (D) Western blot analysis of biotin RNA–hnRNP A1 protein pull down using a goat polyclonal antibody. The sequence of the used RNA oligonucleotides encompassing FANCM positions r.5779_5804 is reported, with the r.5791C>U mutation in bold red, the opal codon enlightened in light grey and the predicted hnRNP A1 binding site created by the mutation boxed in red. As a control for the pull-down efficiency and specificity, we used an antibody against the ELAVL1/HuR protein for which a binding site, boxed in green, is predicted in both RNA oligonucleotides. Input, 10% of total HeLa cell line extract used in the pull-down assay. No, no RNA used as a negative control; 1, normal RNA; 2, RNA carrying the r.5791C>U mutation. The results shown here are representative of two independent experiments.
