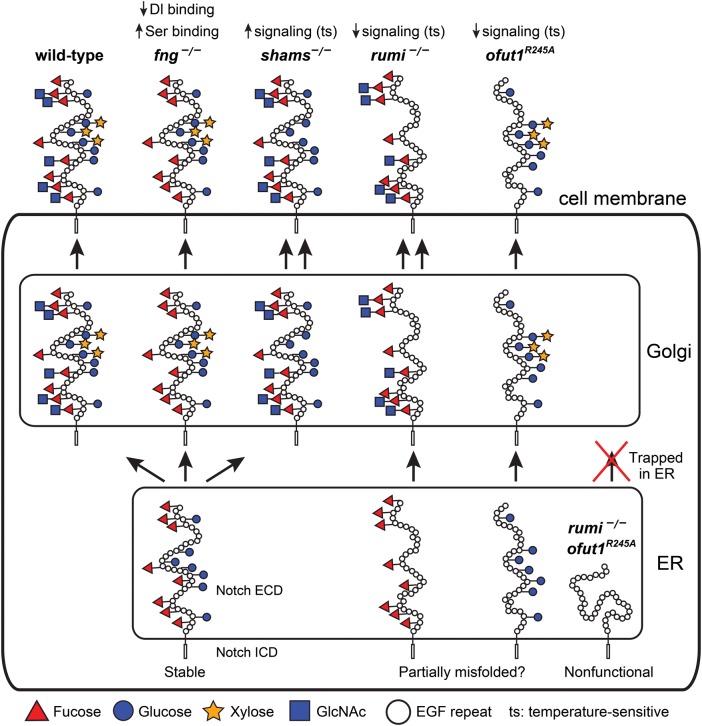
Fig. 2.
Summary of the roles of O-glucose, xylose, O-fucose and GlcNAc in Drosophila Notch signaling. Schematic of the Notch protein in the ER, Golgi and at the cell membrane with its EGF repeat O-glucose and O-fucose glycans are shown in wild-type and various mutant backgrounds. The non-enzymatic chaperone function of Ofut1, which is not reported for its mammalian homologs, is not shown in this figure. Although a non-enzymatic activity has not been formally ruled out for the fly Rumi, O-glucose mutations in Notch recapitulate the rumi loss-of-function phenotypes in the context of Notch signaling. Therefore, rumi mutation is assumed to be equivalent to loss of O-glucose. For simplicity, only EGF repeats are drawn in the extracellular domain, and the intracellular domain is not drawn to scale. The folding of the extracellular domain is arbitrarily drawn. Since Drosophila Notch without O-glucose or O-fucose reaches the cell surface but shows a temperature-sensitive loss of signaling, the extracellular domain of Notch without either of these glycans is drawn as somewhat misfolded. However, other mechanisms might underlie the observed phenotypes. In the absence of both glycans, the Drosophila Notch is trapped in the ER, hence the misfolded schematic. For details, please see the text. It is important to note that the models proposed in this figure are strictly based on Drosophila studies. This figure is available in black and white in print and in color at Glycobiology online.