Abstract
Vibrio species are an abundant and diverse group of bacteria that form associations with phytoplankton. Correlations between Vibrio and phytoplankton abundance have been noted, suggesting that growth is enhanced during algal blooms or that association with phytoplankton provides a refuge from predation. Here, we investigated relationships between particle-associated Vibrio spp. and phytoplankton in Delaware's inland bays (DIB). The relative abundances of particle-associated Vibrio spp. and algal classes that form blooms in DIB (dinoflagellates, diatoms, and raphidophytes) were determined using quantitative PCR. The results demonstrated a significant correlation between particle-associated Vibrio abundance and phytoplankton, with higher correlations to diatoms and raphidophytes than to dinoflagellates. Species-specific associations were examined during a mixed bloom of Heterosigma akashiwo and Fibrocapsa japonica (Raphidophyceae) and indicated a significant positive correlation for particle-associated Vibrio abundance with H. akashiwo but a negative correlation with F. japonica. Changes in Vibrio assemblages during the bloom were evaluated using automated ribosomal intergenic spacer analysis (ARISA), which revealed significant differences between each size fraction but no significant change in Vibrio assemblages over the course of the bloom. Microzooplankton grazing experiments showed that losses of particle-associated Vibrio spp. may be offset by increased growth in the Vibrio population. Moreover, analysis of Vibrio assemblages by ARISA also indicated an increase in the relative abundance for specific members of the Vibrio community despite higher grazing pressure on the particle-associated population as a whole. The results of this investigation demonstrate links between phytoplankton and Vibrio that may lead to predictions of potential health risks and inform future management practices in this region.
INTRODUCTION
Bacteria within the genus Vibrio are naturally abundant in marine and estuarine environments (1), where they exhibit two alternative growth strategies: (i) association with particles as a biofilm or (ii) as free-living bacterioplankton (2). Association with planktonic organisms plays an important role in the ecology of Vibrio (3) by providing an enriched microenvironment for Vibrio spp. (4–6). Previous research demonstrated that planktonic copepods, in particular, enhance the survival and distribution of Vibrio in temperate and tropical areas (7, 8). Associations with plankton may also provide a refuge from grazing by bacterivorous protozoa (9, 10), which can be substantial in some areas (11). Matz et al. (9), for example, showed that the cell density of Vibrio cholerae within a biofilm remained stable in the presence of protozoa, whereas planktonic cells were rapidly eliminated. However, particle association may not always be advantageous, as it may subject the cells to sinking forces or losses through “collateral damage” when host cells are preyed upon (10).
In addition to copepods, Vibrio spp. also form attachments to algal cells (12), and it has been suggested that Vibrio preferentially attaches to algal cells and detritus over whole copepods (8). Increases in Vibrio abundance have been associated with algal blooms and, in some cases, with specific algal groups. For example, Turner et al. (3) showed a significant correlation between phytoplankton abundance and total culturable Vibrio abundance in the coastal waters of Georgia, and high Vibrio abundance has been noted to occur in diatom-dominated phytoplankton assemblages in the Arabian Sea (6). In another study, the abundance of particle-attached Vibrio cholerae increased rapidly (>4 doublings per day) during coastal algal blooms, despite intense predation by protozoa (10). While previous studies have demonstrated a correlation between phytoplankton abundance and Vibrio concentrations in coastal waters, few studies have examined species-specific associations between Vibrio and phytoplankton in the natural environment. In one study, Eiler et al. (13) found a significant correlation between V. cholerae and Prorocentrum spp., suggesting that these interactions may be important determinants regulating intrageneric competition and growth of Vibrio in the marine environment.
Increased occurrences of algal blooms (14) are one consequence of declining coastal water quality in the mid-Atlantic region of the United States (15). Eutrophication of Delaware's inland bays (DIB), which consist of Rehoboth Bay, Indian River Bay, and Little Assawoman Bay, has increased over the last several decades, with high concentrations of nutrient inputs from agricultural and urban sources (16, 17). Several harmful or potentially harmful algal bloom species (HABs) have been identified in DIB, and blooms of harmful algal species occur frequently (18). Among these are several species of harmful dinoflagellates, including Gyrodinium instriatum (19), Karlodinium veneficum (19), and Prorocentrum minimum (20), as well as raphidophytes, a group of species distributed globally in temperate coastal waters and freshwater environments. Marine raphidophytes include genera which are known for fish kills, of which four species bloom annually in DIB: Heterosigma akashiwo, Chattonella subsalsa, Fibrocapsa japonica, and the recently described Viridilobus marinus (19, 21–23).
Although there are extensive data on harmful algal species in DIB, little is known about the ecology of Vibrio in mid-Atlantic estuaries (24) or associations between Vibrio spp. and HABs or other phytoplankton groups in this region. Our goals were to examine community-level and species-specific relationships between particle-associated Vibrio spp. and phytoplankton groups in DIB. Specifically, we examined correlations between the abundance of particle-associated Vibrio spp. and three algal classes—diatoms, dinoflagellates, and raphidophytes—at three sites in DIB over 3 years. We also examined changes in abundance and community composition of particle-associated Vibrio spp. and raphidophytes in size-fractionated water samples during a mixed bloom of Heterosigma akashiwo and Fibrocapsa japonica. In addition, the impacts of microzooplankton grazing on two size fractions of particle-associated and free-living planktonic assemblages of Vibrio were assessed during a separate mixed-raphidophyte bloom. The results of this study are broadly relevant to research on and monitoring of Vibrio and interactions with HABs in the Mid-Atlantic region.
MATERIALS AND METHODS
Field samples.
Water samples were collected weekly from May to September in 2009 to 2011 from three sites within Delaware's inland bays: RB64 in Rehoboth Bay (38°41′59.6″N, 75°06′43.6″W), IR32 in Indian River Bay (38°34′14.5″N, 75°05′04.2″W), and SB10E in Little Assawoman Bay (38°31′15.0″N, 75°03′40.1″W (Fig. 1). Temperature, salinity, and dissolved oxygen (mg liter−1) were measured for each sample using a 556 MPS YSI meter (YSI Inc., Yellow Springs, OH). Dissolved nutrient (NH4, NO3 plus NO2 [NOx], P, and Si) concentrations were determined for water samples collected in 2010 and 2011 only, using a segmented-flow autoanalyzer (Seal Analytical, Mequon, WI) (25, 26). Chlorophyll a (Chl a) concentrations were measured after extraction in 90% acetone (27) on a Turner 10AU fluorometer (Turner Designs, Sunnyvale, CA).
FIG 1.
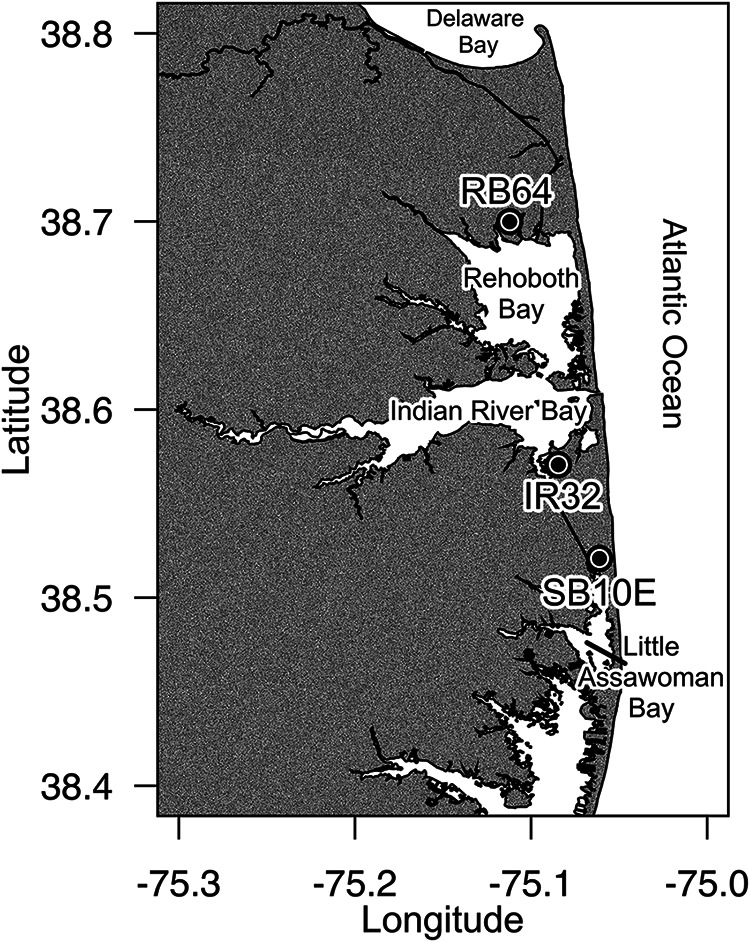
Sampling sites within the Delaware inland bays: Rehoboth Bay at Torquay Canal (RB64), Indian River Bay at Holly Terrace Acres Canal (IR32), and Little Assawoman Bay at South Bethany, Russell Canal East (SB10E). (Map created in R using shoreline data for Delaware and Delaware's inland bays extracted using the NOAA Shoreline Explorer [http://www.ngs.noaa.gov/NSDE/].)
Water samples were filtered under gentle vacuum (∼380 mm Hg) and size fractionated on polycarbonate filters (Millipore Isopore, Billerica, MA) to retain the >3.0-μm size fraction. Filters were immediately placed in CTAB buffer, consisting of 100 mM Tris–HCl (pH 8), 1.4 M NaCl, 2% (wt/vol) cetyltrimethylammonium bromide (CTAB), 0.4% (vol/vol) 2-mercaptoethanol, 1% (wt/vol) polyvinylpyrrolidone, and 20 mM EDTA (28), amended with 20 ng ml−1 pGEM plasmid (Promega, Madison, WI) as an internal standard (21). Filtered samples were stored at −80°C until extraction. Before extraction, all samples were heated at 65°C for 10 min. DNA was extracted as described by Coyne et al. (29) and resuspended in LoTE (3 mM Tris-HCl, 0.2 mM EDTA, pH 7.5) (28, 29). DNA concentrations were determined by spectrophotometry, and samples were diluted to approximately 25 ng μl−1 for molecular analysis.
Primers (Table 1) for the RNA polymerase subunit A (rpoA), a single-copy gene within the Vibrio genome, were previously described by Dalmasso et al. (30). Concentrations of algal class-specific primers (Table 1) targeting the 18S rRNA gene for raphidophytes, dinoflagellates, and diatoms were optimized for quantitative PCR (qPCR) as described by Coyne et al. (21). DNA from size-fractionated water samples was amplified by qPCR in triplicate 10-μl reaction mixtures consisting of 5 μl of SYBR green master mix (Applied Biosystems), 0.9 μM each primer targeting Vibrio rpoA or algal rRNA gene sequences (Table 1), and 1 μl diluted DNA template (∼25 ng). Reaction conditions for amplification of Vibrio rpoA consisted of 2 min at 50°C, 10 min at 95°C, and then 40 cycles at 95°C for 15 s, 53°C for 30 s, and 72°C for 1 min, with added dissociation analysis. Reaction conditions for algal groups were 2 min at 50°C, 10 min at 95°C, and then 40 cycles at 95°C for 15 s, 56°C for 30 s and 72°C for 1 min, with added dissociation analysis. Quantification of the internal pGEM plasmid standard was carried out using a TaqMan-based assay in triplicate 10-μl reaction mixtures consisting of 5 μl of TaqMan universal master mix (Applied Biosystems), 0.9 μM each primer, 0.2 μM TaqMan probe (Table 1), and 1 μl of diluted DNA template. Reaction conditions consisted of 2 min at 50°C, 10 min at 95°C, and then 40 cycles at 95°C for 15 s, 56°C for 30 s and 72°C for 1 min. The relative abundances of Vibrio and algal groups were determined by linear regression analysis using a standard curve generated from rpoA plasmid and 18S rRNA plasmid from each algal group. rpoA and 18S rRNA gene abundances in each sample were normalized to the abundance of pGEM and reported as relative abundance per volume of water filtered.
TABLE 1.
Primer and probe sequences used in this study
| Target (reference) | Primer or probe | Sequence (5′→3′) |
|---|---|---|
| Heterosigma akashiwo 18S rRNA (21) | Hs 1350F | CTAAATAGTGTCGGTAATGCTTCT |
| Hs 1705R | GGCAAGTCACAATAAAGTTCCAT | |
| Hs probe | HEX-CAACGAGTAACGACCTTTGCCGGAA-IBFQ1 | |
| Fibrocapsa japonica 18S rRNA (34) | Fj 1350F | TGCTTTAGTCATTGTGTGCAG |
| Fj 1705R | ACCACAAACTAATGAGGAGGC | |
| Fj probe | FAM-CCCAGGCCTACCGGCCAAGGTTGTA-IBFQ1 | |
| pGEM plasmid DNA (21) | M13F | CCCAGTCACGACGTTGTAAAACG |
| pGEM R | TGTGTGGAATTGTGAGCGGA | |
| pGEM probe | FAM-CACTATAGAATACTCAAGCTTGCATGCCTGCA-IBQF1 | |
| Vibrio sp. rpoA (30) | rpoA 294F | AAATCAGGCTCGGGCCCT |
| rpoA 535R | GCAATTTTRTCDACYGG | |
| Diatom (74) | Diatom 1256F | TAGTGAGGATTGACAGATTGAG |
| Diatom 1536R | CAATAATCTATCCCTATCACGATG | |
| Dinoflagellate (18) | Dino 1662F | CCGATTGAGTGWTCCGGTGAATAA |
| Euk B | GATCCWTCTGCAGGTTCACCTAC | |
| Raphidophyte (75)a | BTG005C | ATCATTACCACACCGATCC |
| Raph-ITS-R | YGCCAGGTGCGTTCGAA | |
| Vibrio sp. ITS (31) | 16S.6 | ACTGGGGTGAAGTCGTAACA |
| 23S.1 | CTTCATCGCCTCTGACTGC |
Modified from reference 75.
Intensive sampling.
Samples were collected during a mixed-raphidophyte bloom of Heterosigma akashiwo (10 to 15 μm in size) and Fibrocapsa japonica (20 to 30 μm in size) at site RB64 on 13 to 16 September 2011. Initial cell concentrations for H. akashiwo were 2.99 × 107 cells liter−1, while F. japonica cell concentrations were 4.1 × 105 cells liter−1. Samples were collected from just below the surface at replicate sites 1 and 2, approximately 10 m apart, at 10:14 a.m. on 13 September 2011 (initial time point, T0) and at 4, 26, 47, and 70 h after T0 (T4, T26, T47, and T70, respectively). Physical parameters and Chl a and nutrient concentrations were measured as described above for each time point. Water was prefiltered on site using a 150-μm filter to remove detritus and zooplankton. Prefiltered samples were then transported to the laboratory and size fractionated onto polycarbonate filters to collect the >20-μm, 3.0- to 20-μm, and 0.2- to 3.0-μm size fractions (which contained the planktonic or “free-living” Vibrio spp.) within 1 h of collection. DNA was extracted from each size fraction as described above. The relative abundances of H. akashiwo (3.0- to 20-μm size fraction) and F. japonica (>20-μm size fraction) were determined using a TaqMan-based assay in triplicate 10-μl reaction mixtures consisting of 5 μl of TaqMan Universal master mix (Applied Biosystems), 0.9 μM species-specific primers (Table 1), 0.2 μM TaqMan probe (Table 1), and 1 μl of diluted DNA template (∼25 ng). Reaction conditions were 2 min at 50°C, 10 min at 95°C, and then 40 cycles at 95°C for 15 s, 56°C for 30 s, and 72°C for 1 min. Quantification of Vibrio spp., dinoflagellates (>20-μm size fraction only), and the pGEM internal standard was carried out as above.
Vibrio assemblages were also evaluated in each size fraction by automated ribosomal intergenic spacer analysis (ARISA) using Vibrio-specific primers (Table 1) (31). The 23S.1 rRNA primer was modified with the fluorescent probe hexachlorofluorescein (HEX) at the 5′ end. ARISA patterns were generated from DNA extracted from known cultured Vibrio species (kindly provided by Gary Richards, USDA ARS, Delaware State University, Dover, DE) for comparison and to identify potential pathogenic species. DNA was amplified after an initial denaturation for 5 min at 94°C for 16 cycles of 94°C for 1 min, 72°C for 1 min (decreased 0.5°C per cycle), and 72°C for 1 min, followed by 22 cycles of 94°C for 1 min, 64°C for 1 min, and 72°C for 1 min. PCR products were then denatured at 94°C for 1 min, followed by a 1-min incubation at 84°C and a 5-min incubation at 72°C. To reduce heteroduplex formation, PCR products were diluted 10-fold in a fresh reaction mixture and subjected to 5 additional cycles of 94°C for 1 min, 64°C for 1 min, and 72°C for 1 min (32). One microliter of the PCR product was combined with 18 μl HiDi formamide (Applied Biosystems) and 1 μl GeneScan-2500 size standard (Applied Biosystems) and denatured at 95°C for 3 min. HEX-labeled PCR products were detected and sized with an ABI Prism 310 genetic analyzer (Applied Biosystems) using GeneScan v. 3.1 software (Applied Biosystems). Results were imported into PeakScanner v.2.0 software (Applied Biosystems) for analysis, and peaks between 50 and 1,000 bases in length were binned at a width of 1 nucleotide (nt). Peaks were aligned and standardized to the total of all peak heights within each sample for comparison between ARISA profiles using T-Rex (33).
Microzooplankton grazing.
Samples were collected from RB64 during a mixed bloom of Heterosigma akashiwo and Fibrocapsa japonica on 23 August and 25 August 2011 for two separate grazing studies. Samples were prefiltered on site through a 150-μm filter to remove zooplankton and detritus. Initial cell concentrations for experiment 1 were 4.3 × 106 cells liter−1 for Heterosigma akashiwo and 2.66 × 104 cells liter−1 for Fibrocapsa japonica. For experiment 2, initial cell concentrations were 1.15 × 107 cells liter−1 and 1.32 × 104 cells liter−1 for H. akashiwo and F. japonica, respectively. For each grazing experiment, an aliquot of the water sample (T0) was size fractionated to achieve >20-μm, 3.0- to 20-μm, and 0.2 to 3.0 μm (free-living) size fractions for both Chl a and qPCR analyses. Site water was filtered through a 0.2-μm filter (Whatman Polycap disposable capsules; GE Healthcare, Piscataway, NJ) for dilution of whole water to achieve dilutions consisting of 25, 50, and 100% of whole water. Diluted samples (n = 4) were enriched with f/2 nutrients. Bottles were incubated at 25°C for 24 h at a light intensity of 228 μmol photons m−2 s−1. Samples were then size fractionated to collect >20-μm, 3.0- to 20-μm, and 0.2- to 3.0-μm (free-living) size fractions for both Chl a and DNA extractions as described above. The relative abundances of H. akashiwo, F. japonica, and Vibrio spp. were determined using qPCR parameters as described above. The Vibrio growth rate (μ) per day was calculated as follows: μ = [ln(relative abundance at T24) − ln(relative abundance at T0)] × day−1. Vibrio apparent growth and grazing rates were calculated by plotting the growth rate of Vibrio versus dilution factor. The negative slope of this relationship is the grazing rate, and the y intercept is the apparent growth rate (34, 35), reported as per day (day−1).
The free-living and particle-associated Vibrio assemblages within each size fraction for the 100% (undiluted) treatments were evaluated by ARISA as described above.
Sequencing.
PCR products generated for ARISA were reamplified using unlabeled 23S.1 and 16S.6 primers (Table 1) as described above. The PCR products were cloned into pCR4-TOPO plasmid vector (Life Technologies, Grand Island, NY, USA). Cloned sequences were then amplified with labeled primers for ARISA to determine the corresponding lengths of each clone. Plasmids with cloned PCR products having lengths of 318, 336, 356, and 412 bp were sequenced bidirectionally using the BigDye Terminator Sequencing kit (Life Technologies, Grand Island, NY).
Statistical analysis.
Statistical comparisons were performed using the R statistical package (36). Relative abundances determined by qPCR were transformed using a square root square root transformation before calculating Pearson's correlations between Vibrio sp. abundance and environmental factors or algal groups. A difference was considered significant if the P value was <0.05. If a significant relationship was detected, a Tukey honestly significant difference (HSD) post hoc test was conducted to determine relationships. Principal-component analysis (PCA) was carried out using the PRIMER 6.1.16 software package (Primer-E, Ivybridge, United Kingdom) to identify factors contributing to differences between sites or years with respect to environmental factors.
Multivariate analysis of ARISA data was carried out using the PRIMER 6.1.16 software package (76, 77). Standardized peak heights from ARISA were subjected to a square root transformation before analysis. The transformed data were then used to produce a Bray-Curtis similarity matrix. Multidimensional scaling (MDS) diagrams were produced from the similarity matrix using the Kruskal fit scheme of 1, with 25 restarts and a minimum stress of 0.01. The analysis of similarity (ANOSIM) function in PRIMER was used to examine relationships between the particle-associated Vibrio community structure in the intensive-sampling and grazing experiments. ANOSIM compares similarities between samples within each group to similarities between groups and generates a value of r between −1 and +1, such that a value of 0 supports the null hypothesis. In the intensive-sampling experiment, ANOSIM was used to test the null hypotheses that Vibrio communities were not significantly different (i) between sites, (ii) between size fractions, or (iii) over time. For the grazing experiments, we used ANOSIM to test the null hypotheses that Vibrio communities were not significantly different (i) between size fractions, (ii) between experiments for each size fraction, or (iii) between T0 and T24 within each size fraction. When ANOSIM revealed a significant difference, Species Contributions to Similarity (SIMPER) was used to identify Vibrio species or operational taxonomic units (OTUs) which contributed to differences in assemblages. The Biota-Environment STepwise (BEST) matching function in PRIMER was conducted to identify environmental variables that were correlated with Vibrio community structure in the intensive-sampling experiment. The BEST analysis calculates the Spearman rank correlation coefficient (ρ) from combinations of variables to find the subset with the highest value of ρ for the Bray-Curtis similarity matrix. For this test, transformed and normalized environmental data were used to construct a Euclidean distance matrix, followed by computation of the rank correlation comparison to the biotic (ARISA) similarity matrix. BEST analysis generated a rank correlation coefficient (rho) such that a value of 0 supports the null hypothesis that the environmental data were not correlated to the biotic data.
Nucleotide sequence accession numbers.
The sequences determined in this study have been submitted to GenBank under accession numbers KT003965 to KT003968.
RESULTS
Field samples.
Altogether, 148 samples were analyzed from three sites in DIB between 2009 and 2011. Temperatures during the collection period ranged from 18.4 to 31.4°C, salinity ranged from 9.3 to 37, and extracted Chl a ranged from 2.82 to 386.8 μg liter−1. The average temperature (24.8°C), salinity (22.9), and Chl a (162.9 mg liter−1) for 2009 were significantly different from those in 2010 (26.7°C, 26.2, and 58.1 mg liter−1) and 2011 (26.5°C, 23.4, and 18.7 mg liter−1) (P < 0.001). The average salinity in 2011 was also significantly lower (23.4) than that in 2010 (26.2) (P < 0.01). Dissolved NOx during the 2010 to 2011 collection period ranged from 0.14 to 49.6 μM, NH4 ranged from 0.3 to 37.5 μM, and PO4 ranged from 0.09 to 2.82 μM. Average concentrations of NOx and PO4 were significantly higher in 2011 (7.4 and 0.7 μM, respectively) than in 2010 (2.7 and 0.4 μM, respectively) (P < 0.001). Principal-component analysis (PCA) showed an increased influence of NOx (PC1, 0.491; PC2, 0.206), N/P ratio (PC1, 0.390; PC2, 0.534), and salinity (PC1, −0.380; PC2, 0.353) on the variability between water samples collected in 2010 versus 2011, with 37.4% variation accounted for on PC1 and 15.3% accounted for on PC2.
Samples that yielded low pGEM values in qPCR analysis were eliminated due to possible inhibition, leaving a total of 132 samples, with 41 samples from IR32, 47 samples from RB64, and 44 samples from SB10E over the 3 years. The relative abundance of particle-associated Vibrio spp. in the >3-μm size fraction as determined by qPCR was significantly correlated to the relative abundance of diatoms and raphidophytes (r = 0.745 and 0.768, respectively; P < 0.001) (Table 2) for all 3 years combined. The correlation between particle-associated Vibrio abundance and dinoflagellates was lower, though still significant (r = 0.560, P < 0.001) (Table 2) for all 3 years combined. Pearson's correlations within collection years showed the highest correlation between particle-associated Vibrio and diatom abundances for 2009 and 2011 (2009, r = 0.764; 2011, r = 0.779), while correlations between Vibrio and raphidophytes were highest in 2010 (r = 0.855) (Table 2). Correlations between particle-associated Vibrio abundance and dinoflagellates were significant only in years 2010 and 2011. Pearson's correlations of particle-associated Vibrio sp. abundance and each phytoplankton group were not significantly different between sites. However, particle-associated Vibrio abundance in the >3-μm size fraction for all 3 years combined was significantly correlated with salinity (r = 0.202, P < 0.05) (Table 2) and Chl a concentrations (r = 0.343, P < 0.001) (Table 2) but not with temperature (Table 2), dissolved oxygen concentrations (data not shown), or nutrient concentrations (data not shown).
TABLE 2.
Correlations between relative abundances of particle-associated Vibrio spp. and algal groups or environmental factors
| Yr | Pearson's correlation coefficienta for correlation between Vibrio abundance and: |
|||||
|---|---|---|---|---|---|---|
| Diatom abundance | Dinoflagellate abundance | Raphidophyte abundance | Temp | Salinity | Chlorophyll a | |
| All | 0.745*** | 0.560*** | 0.768*** | 0.105 | 0.202* | 0.343*** |
| 2009 | 0.764*** | 0.231 | 0.513** | 0.258 | 0.135 | 0.535* |
| 2010 | 0.657*** | 0.550*** | 0.855*** | 0.074 | 0.293* | 0.646*** |
| 2011 | 0.779*** | 0.760*** | 0.560*** | −0.051 | −0.121 | 0.241 |
Significance levels: *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Intensive sampling.
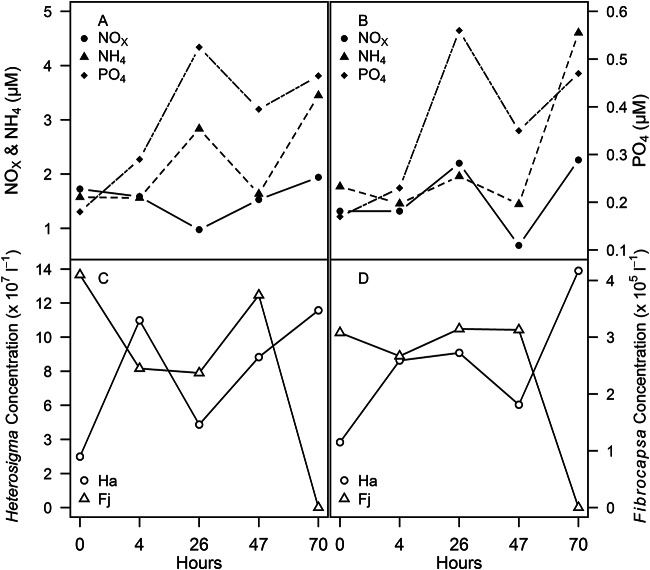
Sampling was conducted on four consecutive days (13 to 16 September 2011) for a total of 10 samples from two replicate sampling locations during a mixed-raphidophyte bloom at RB64. The temperature ranged from 22.9 to 27.2°C, the salinity ranged from 24.2 to 24.8, and the dissolved oxygen concentration ranged from 0.74 to 2.41 mg liter−1. Chl a in the >20-μm fraction ranged from 12.8 to 30.0 μg liter−1, while that in the 3.0- to 20-μm fraction ranged from 11.4 to 21.6 μg liter−1. Dissolved nutrients ranged from 0.69 to 2.3 μM NOx, 1.45 to 4.61 μM NH4, and 0.17 to 0.56 μM PO4 (Fig. 2). Cell counts ranged from 2.99 × 107 to 1.39 × 108 cells liter−1 for Heterosigma akashiwo and from 2.37 × 105 to 4.1 × 105 cells liter−1 for Fibrocapsa japonica (Fig. 2). A significant correlation (Table 3) was identified between particle-associated Vibrio and H. akashiwo abundances in the 3.0- to 20-μm fraction for combined replicate samples (r = 0.788, P < 0.001). Dinoflagellates (>20 μm) were also observed microscopically during the mixed-raphidophyte bloom, and the particle-associated Vibrio abundance in the >20-μm size fraction was more highly correlated with dinoflagellate abundance than with F. japonica abundance. Pearson's correlations between particle-associated Vibrio abundance in the >20-μm fraction and environmental factors were significantly positive and negative for temperature and salinity, respectively (Table 3). However, neither temperature nor salinity was significantly correlated to the 3.0- to 20-μm fraction of particle-associated Vibrio abundance (Table 3).
FIG 2.
Environmental parameters during intensive sampling of a mixed bloom at RB64. (A) Nutrient concentrations (μM) for site 1; (B) nutrient concentrations (μM) for site 2; (C) cell concentrations (cells liter−1) for Heterosigma akashiwo (circles) and Fibrocapsa japonica (triangles) for site 1; (D) cell concentrations (cells liter−1) for Heterosigma akashiwo (circles) and Fibrocapsa japonica (triangles) for site 2.
TABLE 3.
Correlations between relative abundances of particle-associated Vibrio spp. and Heterosigma akashiwo, Fibrocapsa japonica, dinoflagellates, or environmental factors during the intensive-sampling experiment
| Size fraction | Pearson's correlation coefficienta for correlation between Vibrio abundance and: |
||||
|---|---|---|---|---|---|
| Heterosigma abundance | Fibrocapsa abundance | Dinoflagellate abundance | Temp | Salinity | |
| 3.0–20 μm | 0.788*** | 0.015 | −0.133 | ||
| >20 μm | −0.212 | 0.543* | 0.541* | −0.463* | |
Significance levels: *, P < 0.05; ***, P < 0.001.
Analysis of Vibrio community structure during intensive sampling.
Changes in Vibrio assemblages in each size fraction at sites 1 and 2 were examined using ARISA. ARISA of amplified DNA isolated from Vibrio cultures showed major peaks for Vibrio parahaemolyticus at 389 bp, V. tubiashii at 412 bp, V. cholerae at 542 bp, and V. vulnificus at 396 bp. Other constituents of the Vibrio community (OTUs at 318, 336, 356, and 412 bp) were sequenced. BLAST results indicated no significant similarities for OTUs at 318, 336, or 356 bp to Vibrio sequences in GenBank. The sequence for the OTU at 412 bp was 98% identical (100% coverage) to V. corallilyticus strain RE98, a pathogen known to cause mortality in shellfish larvae (37).
Relationships between Vibrio assemblages were investigated by multivariate analysis based on Bray-Curtis similarity matrices implemented in PRIMER, and trends were visualized by construction of a nonmetric multidimensional scaling (MDS) ordination plot from the matrix (Fig. 3). Cluster analysis demonstrated a higher level of similarity in Vibrio community structure within the 0.2- to 3.0-μm size fraction, which contained planktonic or “free-living” Vibrio assemblages (79.71% similar), and in the 3.0- to 20-μm size fraction (67.96% similar) compared to the >20-μm size fraction (28.98% similarity). Statistical analysis by ANOSIM showed that the Vibrio assemblages were not significantly different between sites or over time but that there was a significant difference in assemblages between size fractions (r = 0.571, P = 0.001). SIMPER analyses indicated average dissimilarities of 28.73, 62.07, and 47.66% between assemblages in the free-living and 3.0- to 20-μm fractions, free-living and >20-μm fractions, and 3.0- to 20-μm and >20-μm fractions, respectively. The relative abundance of the OTU at 389 bp, identified as V. parahaemolyticus, was greater in particle-associated fractions and contributed most to the dissimilarity between the Vibrio assemblages in the free-living fraction and the 3.0- to 20-μm fraction (28.37%) and the >20-μm fraction (15.49%). The OTU at 311 bp contributed 14.53% of the dissimilarity between particle-associated Vibrio size fractions where the average relative abundance was lower in the >20-μm size fraction. However, we were not able to isolate this OTU for identification.
FIG 3.
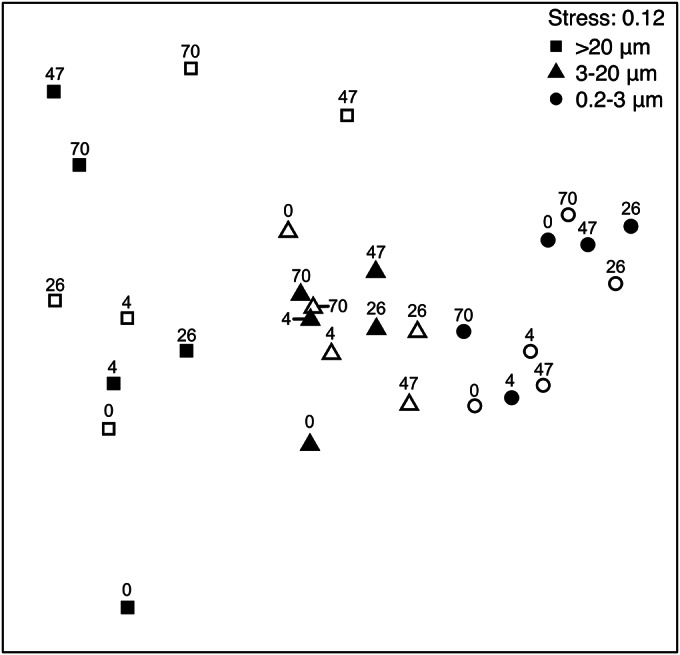
MDS plot of ARISA of Vibrio assemblages from the intensive-sampling experiment during a mixed-raphidophyte bloom in September 2011. Symbols are labeled with the collection time in hours. Open symbols, site 1; closed symbols, site 2.
Microzooplankton grazing.
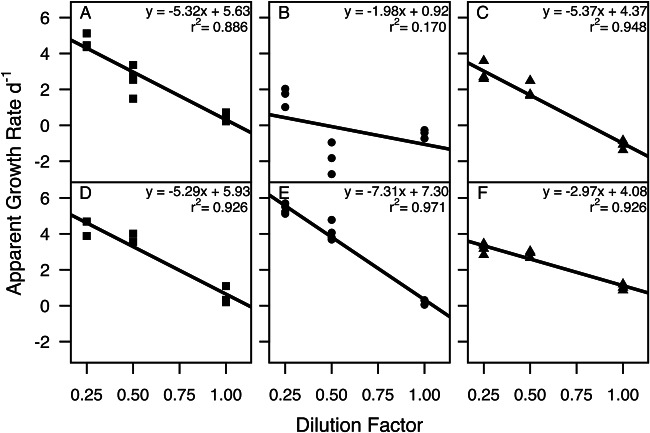
Two grazing experiments were conducted on consecutive days during a mixed bloom of Heterosigma akashiwo and Fibrocapsa japonica in August 2011. For both grazing experiments, environmental parameters at the collection site were similar with regard to temperature and salinity; however, dissolved oxygen was lower at collection time for experiment 2 (Table 4). Chl a concentrations (Table 4) at the start of experiment 1 (T0) were 27.6 μg liter−1 for the >20-μm size fraction, 19.2 μg liter−1 for the 3.0- to 20-μm size fraction, and 9.2 μg liter−1 for the 0.2- to 3.0-μm (free-living) size fraction. In experiment 1, grazing rates on Vibrio, as determined by qPCR, ranged from 1.98 to 5.37 day−1 and were not significantly different between size fractions (P > 0.05) (Fig. 4A to C; Table 5). After 24 h, the relative abundance of Fibrocapsa japonica increased by 155%, whereas Heterosigma akashiwo decreased in relative abundance by 43%, as determined by qPCR. Grazing rates on the total phytoplankton community, based on Chl a concentrations, ranged from −0.036 day−1 in the >20-μm size fraction to 0.337 day−1 in the free-living size fraction for experiment 1 and were significantly higher for the free-living fraction than for the >20-μm and 3.0- to 20-μm size fractions (P < 0.05 and P < 0.0001, respectively) (Table 5).
TABLE 4.
Environmental parameters for grazing experiments at collection time
| Expt | Temp (°C) | Salinity | Dissolved oxygen |
Chlorophyll a (μg liter−1) in size fraction: |
|||
|---|---|---|---|---|---|---|---|
| mg liter−1 | % | >20 μm | 3.0–20.0 μm | Free-living | |||
| 1 | 29.09 | 26.77 | 6.64 | 101.3 | 27.6 | 19.2 | 9.2 |
| 2 | 27.03 | 27.06 | 2.85 | 43 | 31.9 | 9.1 | 10.8 |
FIG 4.
Microzooplankton grazing on size-fractionated Vibrio during a mixed-raphidophyte bloom. (A to C) Experiment 1; (D to F) experiment 2. (A and D) >20-μm size fraction; (B and E) 3.0- to 20-μm size fraction; (C and F) 0.2- to 3.0-μm (free-living) size fraction.
TABLE 5.
Grazing rate and apparent growth (day−1) for grazing experiments
| Expt | Size fraction |
Vibrioa |
Chl aa |
||
|---|---|---|---|---|---|
| r2 | Grazing rate (apparent growth), day−1 | r2 | Grazing rate (apparent growth), day−1 | ||
| 1 | Free-living | 0.886 | 5.37 (4.37) | 0.234 | 0.337 (0.333) |
| 3.0–20 μm | 0.170 | 1.98 (0.92) | 0.762 | 0.248 (0.421)* | |
| >20 μm | 0.948 | 5.32 (5.63) | 0.512 | 0.036 (0.076)*** | |
| 2 | Free-living | 0.926 | 2.97 (4.08)*** | 0.564 | 0.617 (0.310) |
| 3.0–20 μm | 0.971 | 7.31 (7.30)*** | 0.845 | 0.659 (1.219)* | |
| >20 μm | 0.926 | 5.29 (5.93)* | 0.842 | 0.240 (−0.301)** | |
Significance levels: *, P < 0.05; **, P < 0.01; ***, P < 0.001.
In experiment 2, Chl a concentrations at T0 were 31.9 μg liter−1 for the >20-μm size fraction, 9.1 μg liter−1 for the 3.0- to 20-μm size fraction, and 10.8 μg liter−1 for the free-living size fraction (Table 4). Grazing rates on Vibrio during this experiment were significantly different between size fractions, with the highest predation on Vibrio in the 3.0- to 20-μm size fraction and lowest in the free-living size fraction (P < 0.05) (Fig. 4D to F; Table 5). After 24 h, the relative abundance of H. akashiwo increased by 126%, while that of F. japonica decreased by 5% as determined by qPCR. Grazing rates on the total phytoplankton community for experiment 2 were significantly lower in the >20-μm size fraction than in the free-living and 3.0- to 20-μm size fractions (P < 0.05 and P < 0.01, respectively) (Table 5).
When comparing experiments 1 and 2, grazing rates on Vibrio were significantly lower in experiment 2 for the free-living size fraction (P < 0.001) and significantly higher in experiment 2 for the 3.0- to 20-μm fraction (P < 0.01), while grazing rates between the >20-μm fractions were not significantly different. The relative abundance of H. akashiwo was 268% greater at the beginning of experiment 2 compared to experiment 1, while the relative abundance of F. japonica was 201% greater in experiment 1 compared to experiment 2. For the total phytoplankton community, grazing rates were significantly higher in experiment 2 for the >20-μm and 3.0- to 20-μm size fractions compared to experiment 1 (P < 0.001) (Table 5).
Analysis of Vibrio community structure during the grazing experiments.
ARISA was used to examine the changes in the Vibrio community structure during the two grazing experiments. ARISA identified a total of 23 distinct OTUs for both experiments. Analysis of the Vibrio assemblages in experiment 1 by ANOSIM indicated a significant difference between the free-living size fraction and particle-associated Vibrio spp. in both the 3.0- to 20-μm and >20-μm size fractions (r = 0.544 [P = 0.008] and r = 0.492 [P = 0.016], respectively) at 24 h (T24). However, there was no significant difference between particle-associated Vibrio assemblages in the 3.0- to 20-μm and >20-μm size fractions. SIMPER analysis indicated that the OTU at 336 bp contributed the greatest dissimilarity between the free-living and both the 3.0- to 20-μm size fraction (21.63% dissimilarity) and the >20-μm size fraction (20.35% dissimilarity), with greatest abundance in the free-living size fraction. For experiment 2, there was also a significant difference in Vibrio community structure between the free-living and both the 3.0- to 20-μm and >20-μm size fractions (r = 0.214 [P = 0.048] and r= 0.260 [P = 0.024], respectively) and also between the 3.0- to 20-μm and >20-μm size fractions (r = 0.364, P = 0.04). The OTU at 318 bp contributed the most dissimilarity between the free-living size fraction, where it was not detected, and the 3.0- to 20-μm size fraction (15.05% dissimilarity). This OTU also contributed the most dissimilarity between the 3.0- to 20-μm and >20-μm size fractions (14.71% dissimilarity), with higher abundance in the 3.0- to 20-μm size fraction. The OTU at 389 bp, tentatively identified as V. parahaemolyticus, contributed 12.82% to the dissimilarity between particle-associated assemblages and 11.92% to the dissimilarity between free-living and 3.0- to 20-μm size fractions, with greatest abundance in the 3.0- to 20-μm size fraction.
ANOSIM analysis of Vibrio assemblages revealed a significant difference between experiments 1 and 2 for all size fractions combined (r = 0.161, P = 0.005). However, when comparing Vibrio assemblages within each size fraction between these two experiments, there was a significant difference in Vibrio assemblages only in the 3.0- to 20-μm size fractions (r = 0.719, P = 0.029). The OTU at 389 bp, identified as V. parahaemolyticus, contributed the most (15.15%) dissimilarity in this size fraction between experiment 1 (where it was undetected) and experiment 2. Despite significant differences in Vibrio community structure between the 3.0- to 20-μm size fractions in experiments 1 and 2, similarities included the OTU at 318 bp, which contributed 24.52% of the similarity between Vibrio assemblages in this size fraction.
Changes in Vibrio assemblages over time.
There were no significant differences in the Vibrio assemblages between the T0 and T24 time points for the free-living and 3.0- to 20-μm size fractions. In the >20-μm size fraction, however, there was a significant difference in the Vibrio assemblages between T0 and T24 (r = 0.513, P = 0.044). The OTU at 318 bp contributed the most (13.48%) dissimilarity between T0 and T24 in the >20-μm size fraction, where it was undetected after 24 h for both experiments 1 and 2 combined.
Importantly, there were also some trends in the changes in abundance of several OTUs that were consistent for both experiments (Table 6). After 24 h of incubation (T0 to T24), the OTUs at 356, 425, and 542 bp (tentatively identified as V. cholerae) decreased in abundance in the 3.0- to 20-μm size fraction in both experiments. In the >20-μm size fraction, the OTUs at 356 and 318 bp decreased in abundance after 24 h of incubation, while the OTUs at 557 and 412 bp (tentatively identified as V. corallilyticus) increased in abundance for both experiments at T24. Furthermore, the OTU at 318 bp increased in abundance in the 3.0- to 20-μm size fraction in both experiments, where it was undetected at T0, while at the same time it decreased in the >20-μm size fraction in both experiments, where it was undetected at T24.
TABLE 6.
Relative changes in OTU abundance after 24 h that were consistent for both grazing experiments
| OTU (bp) | Change in abundance in size fraction: |
||
|---|---|---|---|
| Free-living | 3.0–20 μm | >20 μm | |
| 318 | NCa | + | − |
| 356 | NC | − | − |
| 412 | NC | NC | + |
| 425 | NC | − | NC |
| 542 | NC | − | NC |
| 557 | NC | NC | + |
NC, no change.
DISCUSSION
In the marine environment, bacteria that are associated with particles represent approximately 10% of the total community (38) but may account for up to 90% of total bacterial production during blooms of phytoplankton (39). Although there is a distinct correlation between bacterial and phytoplankton biomass, little is known about how these communities interact at the species level (40). Several studies suggest that species-specific interactions with bacteria may play a major role in controlling phytoplankton population dynamics (40, 41). For example, bacterial attachment has been shown to stimulate the growth of some dinoflagellates, such as Gambierdiscus toxicus (42), Alexandrium fundyense (43), and Pfiesteria spp. (44), while other bacterial species have been shown to have algicidal effects on phytoplankton (45–47). Furthermore, survival, growth, and abundance of particle-attached bacterioplankton, such as Vibrio, may be positively affected by the release of bioavailable dissolved organic matter (DOM) from phytoplankton (13, 48). Increased growth of pathogenic members of the bacterioplankton due to organic matter from phytoplankton may have serious implications for human and ecosystem health. Previous studies, for example, showed that the growth of V. cholerae increased with amendment of phytoplankton-derived DOM to levels that were 3 orders of magnitude higher than an infectious dose (49).
The objectives of this study were to examine the community-level and species-specific relationships between particle-associated Vibrio and phytoplankton populations in Delaware's inland bays, the influence of environmental factors on these relationships, and the role of microzooplankton grazing in structuring particle-associated Vibrio assemblages. In previous studies, environmental factors such as temperature (4, 48, 50–52) and salinity (51–54) explained the majority of variance in total (free-living and particle-associated) Vibrio abundance compared to other physical parameters (reviewed in references 55 and 56). Temperature and salinity can also influence the formation of Vibrio biofilms in estuarine environments (55, 57). Temperature, in general, is correlated with increased attachment of Vibrio spp., but other, unidentified environmental factors may play a larger role in biofilm formation (reviewed in reference 58). In the study presented here, water temperatures in DIB were not significantly correlated to abundances of particle-associated Vibrio spp. (r = 0.105) (Table 2) for any of the years tested but were significantly correlated to particle-associated Vibrio spp. in the >20-μm size fraction during the intensive-sampling experiment. The relationship between salinity and biofilm formation by Vibrio is more variable and may be species specific, but it can also be influenced by substrate or environmental conditions. The optimal salinity for attachment of V. cholerae to macroalgae and seagrasses, for example, was found to be 1.0 to 1.5% NaCl (12), while others (59) found that changes in salinity had no effect on attachment of this species to chitin. Within DIB, salinity can vary widely and is affected by tidal cycles, evaporation, and rainfall (60). The average salinity was higher in 2010 with a lower range of salinity distributions than in other years during this study, and this may have contributed to the higher relative abundance of particle-associated Vibrio spp. in this year compared to 2009 or 2011 (r = 0.293) (Table 2). This is in contrast to a report by Hsieh et al. (61), who found an increase in particle-associated Vibrio abundance with decreased salinity, and may be due to species-specific interactions between Vibrio and phytoplankton. Indeed, when we measured Vibrio abundance in the intensive-sampling experiment, the particle-associated Vibrio abundance in the >20-μm size fraction (but not the 3.0- to 20-μm fraction) was negatively correlated to salinity.
At the phytoplankton community level, the abundance of particle-associated Vibrio spp. in the >3-μm size fraction was significantly correlated with both diatom and raphidophyte abundances in samples collected in all 3 years (Table 2), while correlations with dinoflagellate abundance were significant only in years 2010 and 2011. With the exception of the 2009 dinoflagellate-Vibrio correlation, the abundance of particle-associated Vibrio spp. was more highly correlated to abundance of each phytoplankton class than to any of the environmental parameters collected. Group- or species-specific associations between Vibrio spp. and phytoplankton may be due to production of algal exudates which activate biofilm formation in Vibrio. Mannitol, for example, is a common exudate from marine phytoplankton (62) and has been shown to induce transcription of the biofilm matrix genes in V. cholerae (63). Other exudates may be involved in species-specific interactions. Chitin, produced as a component of the diatom frustule in some species (64), has been shown to stimulate expression of functional type IV pili in V. parahaemolyticus, resulting in an increase in adherence (64). In addition, Seymour et al. (65) demonstrated a positive chemotactic response by V. alginolyticus to exudates from laboratory cultures of Heterosigma akashiwo, while other bacterial species tested showed no response. Other studies have investigated antagonistic interactions between Vibrio and phytoplankton species in laboratory culture. For example, algicidal activity by a South Carolina isolate of Vibrio spp. resulted in cell lysis of Chattonella subsalsa, Fibrocapsa japonica, and H. akashiwo (66). In another study, Kim et al. (67) demonstrated that the raphidophyte Olisthodiscus luteus (later changed to Heterosigma akashiwo) inhibited the growth of V. alginolyticus by reactive oxygen species-mediated processes. In addition, the bioluminescence of V. fischeri was inhibited 5-fold by the cellular exudate of F. japonica (68). While these laboratory culture experiments suggest a mechanism for species-specific associations, they do not provide much information about the role of these interactions in structuring Vibrio assemblages in the natural environment.
We investigated species-specific associations between Vibrio and two raphidophytes, F. japonica and H. akashiwo, during mixed blooms of these species in DIB. These raphidophytes and their associated Vibrio assemblages were separated by size fractionation, with H. akashiwo retained in the 3- to 20-μm fraction and F. japonica in the >20-μm fraction. Cross-contamination between size fractions was minimal as verified by PCR analysis. The results of our intensive-sampling experiment, in which samples were collected from two replicate locations over the course of 4 days, demonstrated a significant positive correlation between the abundances of particle-associated Vibrio spp. and H. akashiwo and a nonsignificant negative correlation between particle-associated Vibrio and F. japonica abundances (r = −0.212). Changes in the particle-associated Vibrio assemblages during the intensive-sampling experiment were then evaluated using ARISA. ARISA patterns are generated due to heterogeneity of the Vibrio internal transcribed spacer (ITS) length, allowing for discrimination of Vibrio strains (31), but it should be noted that heterogeneity in sequence lengths between strains of the same species may increase the complexity of mixed-community analysis (70). In this study, ANOSIM analysis of ARISA patterns produced by the Vibrio community showed a significant difference in Vibrio population structure associated with each size fraction, supporting the hypothesis that associations between Vibrio spp. and phytoplankton are species specific. Interestingly, ARISA indicated that the relative abundance of the OTU at 389 bp, identified as V. parahaemolyticus, was greater in the both particle-associated size fractions than in the free-living fraction. There are no previous reports of associations between V. parahaemolyticus and raphidophytes, but others have noted higher abundances of this species attached to particles (71).
Attachment of Vibrio to particles, specifically algal cells, in the marine environment may provide a refuge from predation (10). Here, we extended this hypothesis to investigate the effects of grazing on Vibrio assemblages associated with size-fractionated phytoplankton populations during a mixed-raphidophyte bloom. Viral lysis may also increase mortality or impact the bacterial community structure (72), but the effects of viral lysis can be highly variable (see, e.g., reference 73) and were not examined here. Overall, our results indicated that losses to grazing in the particle-associated Vibrio population may be equal to or even greater than losses in the free-living population (Table 5). However, growth rates of the particle-associated Vibrio population were consistently higher in the >20-μm size fraction than in the free-living population, so that in experiment 1, at least, the increased growth conferred by this association may outweigh the losses attributable to predation. From the data collected here, we were not able to identify phytoplankton in the >20-μm size fraction that were associated with Vibrio. Demir et al. (34) found little grazing on F. japonica in DIB, suggesting that association with this species may provide Vibrio with a refuge from predation. However, growth and grazing on Vibrio were not correlated to the relative abundance of F. japonica, which increased in abundance (155%) at T24 in experiment 1 but decreased (by 5%) in experiment 2. Results of the grazing experiments along with the intensive-sampling experiment suggest that the abundance of F. japonica has no impact on growth or grazing of the >20-μm size fraction of particle-associated Vibrio spp. in the natural environment.
In contrast, both growth and grazing rates significantly increased for Vibrio in the 3.0- to 20-μm size fraction between experiments 1 and 2 (Fig. 4B and E), corresponding to an increase (119%) in the relative abundance of Heterosigma akashiwo in this size fraction between experiments. Demir et al. (34) showed higher microzooplankton grazing pressure on H. akashiwo from DIB compared to other raphidophyte species, implying that Vibrio spp. that are associated with Heterosigma will also face increased predation. It is notable, though, that growth rates for Vibrio were also highest for this size fraction in experiment 2. These results support the idea that Vibrio assemblages exhibit multiple growth strategies, as suggested by Worden et al. (10), where free-living Vibrio may grow rapidly (Fig. 4C and F) with nutrient input, but that association with particles in the environment can also be beneficial, resulting in an increased growth rate (Fig. 4A, D, and E) despite increased grazing pressure.
The effects of grazing on the population structure of free-living and particle-associated Vibrio spp. revealed differential impacts on Vibrio species, such that association with particles may confer protection for some species over others. Furthermore, several species or OTUs, as noted above, consistently increased or decreased within the same size fraction for both grazing experiments (Table 6), supporting the hypothesis that associations between Vibrio and phytoplankton are species specific. There was also evidence that some particle-associated Vibrio species (the OTU at 318 bp, for example) may increase in abundance in one size fraction while decreasing in other size fractions. It is possible that Vibrio species remained within the same size fraction for the duration of the experiment, in which case specific association with phytoplankton that are more heavily grazed within one size fraction would result in a decrease in their abundance, while associations with other size fractions confers protection for the same species. Alternatively, a concurrent increase of some OTUs within one size fraction and a decrease in another may be due to a remobilization of Vibrio cells from one size fraction to another to avoid predation. In either case, the results of our study indicate that species-specific associations between Vibrio and phytoplankton provide some species with a clear advantage in periods of high grazing pressure.
Enhanced growth of Vibrio pathogens even in the presence of increased grazing pressure may have significant implications for human and ecosystem health. The OTU at 389 bp, for example, was tentatively identified as V. parahaemolyticus, a human pathogen responsible for gastroenteritis and wound infection (24, 56). This OTU was a large portion of the free-living size fraction at the beginning of the grazing in experiment 2 but decreased in relative abundance in this fraction while increasing in the 3.0- to 20-μm and >20-μm size fractions. In a similar manner, the OTU at 412 bp, tentatively identified as V. corallilyticus, an oyster pathogen (37), increased in the >20-μm size fraction in both grazing experiments. These results suggest that association with particles may provide some Vibrio species, including potential pathogens, with a growth advantage over other members of the community, in spite of increased grazing pressure on the population as a whole.
The results of this investigation may lead to predictions of potential outbreaks of Vibrio by association with algal bloom species. Distinct differences between Vibrio assemblages associated with different size fractions of phytoplankton in DIB suggest not only that blooms are a vector for Vibrio but that species-specific associations may determine the potential risk to human and ecosystem health. Specifically, our investigation points to associations that favor interactions between Vibrio and H. akashiwo over F. japonica, while results of microzooplankton grazing experiments also demonstrate that these associations may benefit some species of Vibrio while at the same time result in greater loss to the population as a whole due to grazing.
ACKNOWLEDGMENTS
This work was supported by Delaware Sea Grant (grant R/HCE-4 to KJC), the National Oceanic and Atmospheric Association (NOAA) Monitoring and Event Response for Harmful Algal Blooms (MERHAB) program (grant NA10NOS4780141 to K.J.C. and Dianne I. Greenfield, contribution number 184), and the Experimental Program to Stimulate Competitive Research (EPSCoR) (grant EPS-0814251 to K.J.C. and E. Fidelma Boyd).
We thank E. Fidelma Boyd and the University of Delaware Citizen Monitoring Program for providing samples for analysis.
REFERENCES
- 1.Grimes DJ. 1991. Ecology of estuarine bacteria capable of causing human disease: a review. Estuaries Coasts 14:345–360. doi: 10.2307/1352260. [DOI] [Google Scholar]
- 2.Yildiz FH, Visick KL. 2009. Vibrio biofilms: so much the same yet so different. Trends Microbiol 17:109–118. doi: 10.1016/j.tim.2008.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Turner JW, Good B, Cole D, Lipp EK. 2009. Plankton composition and environmental factors contribute to Vibrio seasonality. ISME J 3:1082–1092. doi: 10.1038/ismej.2009.50. [DOI] [PubMed] [Google Scholar]
- 4.Huq A, Sack RB, Nizam A, Longini IM, Nair GB, Ali A, Morris JG, Khan MNH, Siddique AK, Yunus M, Albert MJ, Sack DA, Colwell RR. 2005. Critical factors influencing the occurrence of Vibrio cholerae in the environment of Bangladesh. Appl Environ Microbiol 71:4645–4654. doi: 10.1128/AEM.71.8.4645-4654.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eiler A, Bertilsson S. 2006. Detection and quantification of Vibrio populations using denaturant gradient gel electrophoresis. J Microbiol Methods 67:339–348. doi: 10.1016/j.mimet.2006.04.002. [DOI] [PubMed] [Google Scholar]
- 6.Asplund ME, Rehnstam-Holm A-S, Atnur V, Raghunath P, Saravanan V, Härnström K, Collin B, Karunasagar I, Godhe A. 2011. Water column dynamics of Vibrio in relation to phytoplankton community composition and environmental conditions in a tropical coastal area. Environ Microbiol 13:2738–2751. doi: 10.1111/j.1462-2920.2011.02545.x. [DOI] [PubMed] [Google Scholar]
- 7.Lizárraga-Partida ML, Mendez-Gómez E, Rivas-Montaño AM, Vargas-Hernández E, Portillo-López A, González-Ramírez AR, Huq A, Colwell RR. 2009. Association of Vibrio cholerae with plankton in coastal areas of Mexico. Environ Microbiol 11:201–208. doi: 10.1111/j.1462-2920.2008.01753.x. [DOI] [PubMed] [Google Scholar]
- 8.Tamplin ML, Gauzens AL, Huq A, Sack DA, Colwell RR. 1990. Attachment of Vibrio cholerae serogroup O1 to zooplankton and phytoplankton of Bangladesh waters. Appl Environ Microbiol 56:1977–1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Matz C, McDougald D, Moreno AM, Yung PY, Yildiz FH, Kjelleberg S. 2005. Biofilm formation and phenotypic variation enhance predation-driven persistence of Vibrio cholerae. Proc Natl Acad Sci U S A 102:16819–16824. doi: 10.1073/pnas.0505350102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Worden AZ, Seidel M, Smriga S, Wick A, Malfatti F, Bartlett DH, Azam F. 2006. Trophic regulation of Vibrio cholerae in coastal marine waters. Environ Microbiol 8:21–29. doi: 10.1111/j.1462-2920.2005.00863.x. [DOI] [PubMed] [Google Scholar]
- 11.Beardsley C, Pernthaler J, Wosniok W, Amann R. 2003. Are readily culturable bacteria in coastal North Sea waters supressed by selective grazing mortality? Appl Environ Microbiol 69:2624–2630. doi: 10.1128/AEM.69.5.2624-2630.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hood MA, Winter PA. 1997. Attachment of Vibrio cholerae under various environmental conditions and to selected substrates. FEMS Microbiol Ecol 22:215–223. doi: 10.1111/j.1574-6941.1997.tb00373.x. [DOI] [Google Scholar]
- 13.Eiler A, Johansson M, Bertilsson S. 2006. Environmental influences on Vibrio populations in northern temperate and boreal coastal waters (Baltic and Skagerrak Seas). Appl Environ Microbiol 72:6004–6011. doi: 10.1128/AEM.00917-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bricker SB, Longstaff B, Dennison W, Jones A, Boicourt K, Wicks C, Woerner J. 2008. Effects of nutrient enrichment in the nation's estuaries: a decade of change. Harmful Algae 8:21–32. doi: 10.1016/j.hal.2008.08.028. [DOI] [Google Scholar]
- 15.Kiddon JA, Paul JF, Buffum HW, Strobel CS, Hale SS, Cobb D, Brown BS. 2003. Ecological condition of US Mid-Atlantic estuaries, 1997-1998. Mar Pollut Bull 46:1224–1244. doi: 10.1016/S0025-326X(03)00322-9. [DOI] [PubMed] [Google Scholar]
- 16.Price K. 1998. A framework for a Delaware Inland Bays environmental classification. Environ Monit Assess 51:285–298. doi: 10.1023/A:1005951706152. [DOI] [Google Scholar]
- 17.Sallade YE, Sims JT. 1997. Phosphorus transformations in the sediments of Delaware's agricultural drainageways. II. Effect of reducing conditions on phosphorus release. J Environ Qual 26:1579. [Google Scholar]
- 18.Handy SM, Demir E, Hutchins DA, Portune KJ, Whereat EB, Hare C, Rose J, Warner M, Farestad M, Cary SC. 2008. Using quantitative real-time PCR to study competition and community dynamics among Delaware Inland Bays harmful algae in field and laboratory studies. Harmful Algae 7:599–613. doi: 10.1016/j.hal.2007.12.018. [DOI] [Google Scholar]
- 19.Whereat EB. 2013. Harmful algae report. The University of Delaware Citizen Monitoring Program, Lewes, DE. [Google Scholar]
- 20.Warner ME, Madden ML. 2007. The impact of shifts to elevated irradiance on the growth and photochemical activity of the harmful algae Chattonella subsalsa and Prorocentrum minimum from Delaware. Harmful Algae 6:332–342. doi: 10.1016/j.hal.2006.04.007. [DOI] [Google Scholar]
- 21.Coyne KJ, Handy SM, Demir E, Whereat EB, Hutchins DA, Portune KJ, Doblin MA, Cary SC. 2005. Improved quantitative real-time PCR assays for enumeration of harmful algal species in field samples using an exogenous DNA reference standard. Limnol Oceanogr Methods 3:381–391. doi: 10.4319/lom.2005.3.381. [DOI] [Google Scholar]
- 22.Handy SM, Hutchins DA, Cary SC, Coyne KJ. 2006. Simultaneous enumeration of multiple raphidophyte species by quantitative real-time PCR: capabilities and limitations. Limnol Oceanogr Methods 4:193–204. doi: 10.4319/lom.2006.4.193. [DOI] [Google Scholar]
- 23.Demir-Hilton E, Hutchins DA, Czymmek KJ, Coyne KJ. 2012. Description of Viridilobus marinus (gen. et sp. nov.), a new raphidophye from Delaware's Inland Bays. J Phycol 48:1220–1231. doi: 10.1111/j.1529-8817.2012.01212.x. [DOI] [PubMed] [Google Scholar]
- 24.Johnson CN, Bowers JC, Griffitt KJ, Molina V, Clostio RW, Pei S, Laws E, Paranjpye RN, Strom MS, Chen A, Hasan NA, Huq A, Noriea NF, Grimes DJ, Colwell RR. 2012. Ecology of Vibrio parahaemolyticus and Vibrio vulnificus in the coastal and estuarine waters of Louisiana, Maryland, Mississippi, and Washington (United States). Appl Environ Microbiol 78:7249–7257. doi: 10.1128/AEM.01296-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johnson K, Petty R. 1983. Determination of nitrate and nitrite in seawater by flow injection analysis. Limnol Oceanogr 28:1260–1266. doi: 10.4319/lo.1983.28.6.1260. [DOI] [Google Scholar]
- 26.Hansen HP, Koroleff F. 1999. Determination of nutrients, p 159–228. In Grasshoff K, Kremling K, Ehrhardt M (ed), Methods of seawater analysis, 3rd ed John Wiley & Sons, Hoboken, NJ. [Google Scholar]
- 27.Welschmeyer NA. 1994. Fluorometric of chlorophyll a in the presence of analysis chlorophyll b and pheopigments. Limnol Oceanogr 39:1985–1992. doi: 10.4319/lo.1994.39.8.1985. [DOI] [Google Scholar]
- 28.Dempster EL, Pryor KV, Francis D, Young JE, Rogers HJ. 1999. Rapid DNA extraction from ferns for PCR-based analyses. Biotechniques 27:66–68. [DOI] [PubMed] [Google Scholar]
- 29.Coyne KJ, Hutchins DA, Hare C, Cary SC. 2001. Assessing temporal and spatial variability in Pfiesteria piscicida distributions using molecular probing techniques. Aquat Microb Ecol 24:275–285. doi: 10.3354/ame024275. [DOI] [Google Scholar]
- 30.Dalmasso A, La Neve F, Suffredini E, Croci L, Serracca L, Bottero MT, Civera T. 2009. Development of a PCR assay targeting the rpoA gene for the screening of Vibrio genus. Food Anal Methods 2:317–324. doi: 10.1007/s12161-009-9089-9. [DOI] [Google Scholar]
- 31.Hoffmann M, Brown EW, Feng PCH, Keys CE, Fischer M, Monday SR. 2010. PCR-based method for targeting 16S-23S rRNA intergenic spacer regions among Vibrio species. BMC Microbiol 10:90. doi: 10.1186/1471-2180-10-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thompson JR, Marcelino LA, Polz MF. 2002. Heteroduplexes in mixed-template amplifications: formation, consequence and elimination by “reconditioning PCR”. Nucleic Acids Res 30:2083–2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Culman SW, Bukowski R, Gauch HG, Cadillo-Quiroz H, Buckley DH. 2009. T-REX: software for the processing and analysis of T-RFLP data. BMC Bioinformatics 10:171. doi: 10.1186/1471-2105-10-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Demir E, Coyne KJ, Doblin MA, Handy SM, Hutchins DA. 2008. Assessment of microzooplankton grazing on Heterosigma akashiwo using a species-specific approach combining quantitative real-time PCR (QPCR) and dilution methods. Microb Ecol 55:583–594. doi: 10.1007/s00248-007-9263-9. [DOI] [PubMed] [Google Scholar]
- 35.Landry MR, Hassett RP. 1982. Estimating the grazing impact of marine micro-zooplankton. Mar Biol 67:283–288. doi: 10.1007/BF00397668. [DOI] [Google Scholar]
- 36.R Core Team. 2012. R: a language and environment for statistical computing, 2.15.2. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- 37.Richards GP, Watson MA, Needleman DS, Church KM, Hase CC. 2015. Mortalities of Eastern and Pacific oyster larvae caused by the pathogens Vibrio coralliilyticus and Vibrio tubiashii. Appl Environ Microbiol 81:292–297. doi: 10.1128/AEM.02930-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Middelboe M, Søndergaard M, Letarte Y, Borch NH. 1995. Attached and free-living bacteria: production and polymer hydrolysis during a diatom bloom. Microb Ecol 29:231–248. doi: 10.1007/BF00164887. [DOI] [PubMed] [Google Scholar]
- 39.Smith DC, Steward GF, Long RA, Azam F. 1995. Bacterial mediation of carbon fluxes during a diatom bloom in a mesocosm. Deep Sea Res II Top Stud Oceanogr 42:75–97. doi: 10.1016/0967-0645(95)00005-B. [DOI] [Google Scholar]
- 40.Rooney-Varga JN, Giewat MW, Savin MC, Sood S, LeGresley M, Martin JL. 2005. Links between phytoplankton and bacterial community dynamics in a coastal marine environment. Microb Ecol 49:163–175. doi: 10.1007/s00248-003-1057-0. [DOI] [PubMed] [Google Scholar]
- 41.Lovejoy C, Bowman JP, Hallegraeff GM. 1998. Algicidal effects of a novel marine Pseudoalteromonas isolate (class Proteobacteria, gamma subdivision) on harmful algal bloom species of the genera Chattonella, Gymnodinium, and Heterosigma. Appl Environ Microbiol 64:2806–2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sakami T, Nakahara H, Chinain M, Ishida Y. 1999. Effects of epiphytic bacteria on the growth of the toxic dinoflagellate Gambierdiscus toxicus (Dinophyceae). J Exp Mar Biol Ecol 233:231–246. doi: 10.1016/S0022-0981(98)00130-0. [DOI] [Google Scholar]
- 43.Ferrier M, Martin JL, Rooney-Varga JN. 2002. Stimulation of Alexandrium fundyense growth by bacterial assemblages from the Bay of Fundy. J Appl Microbiol 92:706–716. doi: 10.1046/j.1365-2672.2002.01576.x. [DOI] [PubMed] [Google Scholar]
- 44.Alavi M, Miller T, Erlandson K, Schneider R, Belas R. 2001. Bacterial community associated with Pfiesteria-like dinoflagellate cultures. Environ Microbiol 3:380–396. doi: 10.1046/j.1462-2920.2001.00207.x. [DOI] [PubMed] [Google Scholar]
- 45.Yang Y, Hu X, Zhang J, Gong Y. 2013. Community level physiological study of algicidal bacteria in the phycospheres of Skeletonema costatum and Scrippsiella trochoidea. Harmful Algae 28:88–96. doi: 10.1016/j.hal.2013.05.015. [DOI] [Google Scholar]
- 46.Skerratt J, Bowman J, Hallegraeff G, James S, Nichols P. 2002. Algicidal bacteria associated with blooms of a toxic dinoflagellate in a temperate Australian estuary. Mar Ecol Prog Ser 244:1–15. doi: 10.3354/meps244001. [DOI] [Google Scholar]
- 47.Mayali X, Azam F. 2004. Algicial bacteria in the sea and their impact on algal blooms. J Eukaryot Microbiol 51:139–144. doi: 10.1111/j.1550-7408.2004.tb00538.x. [DOI] [PubMed] [Google Scholar]
- 48.Eiler A, Gonzalez-Rey C, Allen S, Bertilsson S. 2007. Growth response of Vibrio cholerae and other Vibrio spp. to cyanobacterial dissolved organic matter and temperature in brackish water. FEMS Microbiol Ecol 60:411–418. doi: 10.1111/j.1574-6941.2007.00303.x. [DOI] [PubMed] [Google Scholar]
- 49.Mouriño-Pérez RR, Worden AZ, Azam F. 2003. Growth of Vibrio cholerae O1 in red tide waters off California. Appl Environ Microbiol 69:6923–6931. doi: 10.1128/AEM.69.11.6923-6931.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lobitz B, Beck L, Huq A, Wood B, Fuchs G, Faruque A, Colwell RR. 2000. Climate and infectious disease: use of remote sensing for detection of Vibrio cholerae by indirect measurement. Proc Natl Acad Sci U S A 97:1438. doi: 10.1073/pnas.97.4.1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Singleton FL, Attwell R, Jangi S, Colwell RR. 1982. Effects of temperature and salinity on growth of Vibrio cholerae. Appl Environ Microbiol 44:1047–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Froelich B, Bowen J, Gonzalez R, Snedeker A, Noble R. 2013. Mechanistic and statistical models of total Vibrio abundance in the Neuse River Estuary. Water Res 47:5783–5793. doi: 10.1016/j.watres.2013.06.050. [DOI] [PubMed] [Google Scholar]
- 53.Randa MA, Polz MF, Lim E. 2004. Effects of temperature and salinity on Vibrio vulnificus population dynamics as assessed by quantitative PCR. Appl Environ Microbiol 70:5469. doi: 10.1128/AEM.70.9.5469-5476.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Motes ML, DePaola A, Cook DW, Veazey JE, Hunsucker JC, Garthright WE, Blodgett RJ, Chirtel SJ. 1998. Influence of water temperature and salinity on Vibrio vulnificus in Northern Gulf and Atlantic Coast oysters (Crassostrea virginica). Appl Environ Microbiol 64:1459–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Takemura AF, Chien DM, Polz MF. 2014. Associations and dynamics of Vibrionaceae in the environment, from the genus to the population level. Front Microbiol 5:38. doi: 10.3389/fmicb.2014.00038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Blackwell KD, Oliver JD. 2008. The ecology of Vibrio vulnificus, Vibrio cholerae, and Vibrio parahaemolyticus in North Carolina estuaries. J Microbiol 46:146–153. doi: 10.1007/s12275-007-0216-2. [DOI] [PubMed] [Google Scholar]
- 57.McDougald D, Lin WH, Rice SA, Kjelleberg S. 2006. The role of quorum sensing and the effect of environmental conditions on biofilm formation by strains of Vibrio vulnificus. Biofouling 22:133–144. doi: 10.1080/08927010600691879. [DOI] [PubMed] [Google Scholar]
- 58.Lutz C, Erken M, Noorian P, Sun S, McDougald D. 2013. Environmental reservoirs and mechanisms of persistence of Vibrio cholerae. Front Microbiol 4:375. doi: 10.3389/fmicb.2013.00375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stauder M, Vezzulli L, Pezzati E, Repetto B, Pruzzo C. 2010. Temperature affects Vibrio cholerae O1 El Tor persistence in the aquatic environment via an enhanced expression of GbpA and MSHA adhesins. Environ Microbiol Rep 2:140–144. doi: 10.1111/j.1758-2229.2009.00121.x. [DOI] [PubMed] [Google Scholar]
- 60.Zhang Y, Fu F-X, Whereat EB, Coyne KJ, Hutchins DA. 2006. Bottom-up controls on a mixed-species HAB assemblage: a comparison of sympatric Chattonella subsalsa and Heterosigma akashiwo (Raphidophyceae) isolates from the Delaware Inland Bays, USA. Harmful Algae 5:310–320. doi: 10.1016/j.hal.2005.09.001. [DOI] [Google Scholar]
- 61.Hsieh JL, Fries JS, Noble RT. 2007. Vibrio and phytoplankton dynamics during the summer of 2004 in a eutrophying estuary. Ecol Appl 17:S102–S109. doi: 10.1890/05-1274.1. [DOI] [Google Scholar]
- 62.Dittami SM, Aas HTN, Paulsen BS, Boyen C, Edvardsen B, Tonon T. 2011. Mannitol in six autotrophic stramenopiles and Micromonas. Plant Signal Behav 6:1237–1239. doi: 10.4161/psb.6.8.16404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ymele-Leki P, Houot L, Watnick PI. 2013. Mannitol and the mannitol-specific enzyme IIB subunit active Vibrio cholerae biofilm formation. Appl Environ Microbiol 79:4675–4683. doi: 10.1128/AEM.01184-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Frischkorn KR, Stojanovski A, Paranjpye R. 2013. Vibrio parahaemolyticus type IV pili mediate interactions with diatom-derived chitin and point to an unexplored mechanism of environmental persistence. Environ Microbiol 15:1416–1427. doi: 10.1111/1462-2920.12093. [DOI] [PubMed] [Google Scholar]
- 65.Seymour JR, Ahmed T, Stocker R. 2009. Bacterial chemotaxis towards the extracellular products of the toxic phytoplankton Heterosigma akashiwo. J Plankton Res 31:1557–1561. doi: 10.1093/plankt/fbp093. [DOI] [Google Scholar]
- 66.Liu J, Lewitus AJ, Kempton JW, Wilde SB. 2008. The association of algicidal bacteria and raphidophyte blooms in South Carolina brackish detention ponds. Harmful Algae 7:184–193. doi: 10.1016/j.hal.2007.07.001. [DOI] [Google Scholar]
- 67.Kim D, Nakamura A, Okamoto T, Komatsu N, Oda T, Ishimatsu A, Muramatsu T. 1999. Toxic potential of the raphidophyte Olisthodiscus luteus: mediation by reactive oxygen species. J Plankton Res 21:1017–1027. doi: 10.1093/plankt/21.6.1017. [DOI] [Google Scholar]
- 68.Van Rijssel M, de Boer MK, Tyl MR, Gieskes WWC. 2007. Evidence for inhibition of bacterial luminescence by allelochemicals from Fibrocapsa japonica (Raphidophyceae), and the role of light and microalgal growth rate. Hydrobiologia 596:289–299. [Google Scholar]
- 69.Reference deleted.
- 70.Crosby LD, Criddle CS. 2003. Understanding bias in microbial community analysis techniques due to rrn operon copy number heterogeneity. Biotechniques 34:790–794, 796, 798 passim. [DOI] [PubMed] [Google Scholar]
- 71.Venkateswaran K, Kiiyukia C, Nakanishi K, Nakano H, Matsuda O, Hashimoto H. 1990. The role of sinking particles in the overwintering process of Vibrio parahaemolyticus in a marine environment. FEMS Microbiol Lett 73:159–166. doi: 10.1111/j.1574-6968.1990.tb03936.x. [DOI] [Google Scholar]
- 72.Weinbauer MG, Hornák K, Jezbera J, Nedoma J, Dolan JR, Šimek K. 2007. Synergistic and antagonistic effects of viral lysis and protistan grazing on bacterial biomass, production and diversity. Environ Microbiol 9:777–788. doi: 10.1111/j.1462-2920.2006.01200.x. [DOI] [PubMed] [Google Scholar]
- 73.Horňák K, Mašín M, Jezbera J, Bettarel Y, Nedoma J, Sime-Ngando T, Šimek K. 2005. Effects of decreased resource availability, protozoan grazing and viral impact on a structure of bacterioplankton assemblage in a canyon-shaped reservoir. FEMS Microbiol Ecol 52:315–327. doi: 10.1016/j.femsec.2004.11.013. [DOI] [PubMed] [Google Scholar]
- 74.Tilney CL, Pokrzywinski KL, Coyne KJ, Warner ME. 2014. Effects of a bacterial algicide, IRI-160AA, on dinoflagellates and the microbial community in microcosm experiments. Harmful Algae 39:210–222. doi: 10.1016/j.hal.2014.08.001. [DOI] [Google Scholar]
- 75.Connell LB. 2002. Rapid identificationof marine algae (Raphidophyceae) using three-primer PCR amplification of nuclear internal transcribed spacer (ITS) regions from fresh and archived material. Phycologia 41:15–21. doi: 10.2216/i0031-8884-41-1-15.1. [DOI] [Google Scholar]
- 76.Clarke KR, Gorley RN. 2006. PRIMER v6: user manual/tutorial. PRIMER-E, Plymouth, United Kingdom. [Google Scholar]
- 77.Clarke KR, Warwick RM. 1994. Change in marine communities: an approach to statistical analysis and interpretation. PRIMER-E, Plymouth, United Kingdom. [Google Scholar]