IFN-γ is able to modulate endosome dynamics in myelocytic cells, but the molecular mechanisms behind this process remain to be elucidated. Rab20 is identified as part of the molecular machinery that links immune activation and control of endocytic function in macrophages.
Abstract
Little is known about the molecular players that regulate changes in the endocytic pathway during immune activation. Here we investigate the role of Rab20 in the endocytic pathway during activation of macrophages. Rab20 is associated with endocytic structures, but the function of this Rab GTPase in the endocytic pathway remains poorly characterized. We find that in macrophages, Rab20 expression and endosomal association significantly increase after interferon-γ (IFN-γ) treatment. Moreover, IFN-γ and Rab20 expression induce a dramatic enlargement of endosomes. These enlarged endosomes are the result of homotypic fusion promoted by Rab20 expression. The expression of Rab20 or the dominant-negative mutant Rab20T19N does not affect transferrin or dextran 70 kDa uptake. However, knockdown of Rab20 accelerates epidermal growth factor (EGF) trafficking to LAMP-2–positive compartments and EGF receptor degradation. Thus this work defines a function for Rab20 in the endocytic pathway during immune activation of macrophages.
INTRODUCTION
Interferon-γ (IFN-γ) is the key protective cytokine for macrophages and enhances their antimicrobial capacity and stimulates the secretion of proinflammatory mediators (Schroder et al., 2004). There is substantial evidence that IFN-γ plays diverse roles during modulation of both the endocytic and phagocytic pathways (Barry et al., 2011). For example, IFN-γ induces trafficking of the gp91 and p22 subunits of NADPH oxidase from intracellular compartments to the plasma membrane, correlating with increased reactive oxygen species production (Casbon et al., 2012). Moreover, IFN-γ treatment results in the formation of enlarged endosomes in macrophages (Montaner et al., 1999). IFN-γ also induces an increase in the association of Rab20 with phagosomes (Pei et al., 2014). Although the regulation of intracellular trafficking by IFN-γ is well described, the molecular players that control this process are poorly characterized.
Rab GTPases are central regulators of membrane trafficking and control cargo selection, vesicle budding, motility, tethering, and fusion (Zerial and McBride, 2001; Stenmark, 2009). There are >60 family members in humans, which specifically localize in different intracellular compartments (Stenmark and Olkkonen, 2001). Regulation of the expression of Rab GTPases is emerging as an important mechanism by which intracellular transport is differentially modulated by cytokines (Pei et al., 2012). The expression of Rab20 is up-regulated in macrophages by mycobacterial infection in an NF-κB–dependent manner (Gutierrez et al., 2008). Rab20 expression is also highly increased upon different TLR ligand stimulation in dendritic cells (Torri et al., 2010). However, little is known about the expression of Rab20 during cytokine activation in immune cells.
Although Rab20 has been associated with many components of the endocytic pathway, its precise role in this pathway has not been investigated. Rab20 was first reported to be associated with apical endocytic organelles in mouse kidney proximal tubule cells (Lütcke et al., 1994). Later Rab20 was found to localize to the cis-Golgi and medium compartment of Golgi apparatus in HeLa cells (Amillet et al., 2006) and colocalize with the subunit E of the vacuolar ATPase (v-ATPase) in kidney, suggesting that Rab20 might regulate the trafficking of v-ATPase (Curtis and Gluck, 2005). Rab20 has also been suggested as a putative regulator of connexin 43 trafficking from the perinuclear region to the endoplasmic reticulum (Das Sarma et al., 2008). In addition, Rab20 is transiently associated with macropinosomes (Egami and Araki, 2012b) and phagosomes (Seto et al., 2011; Egami and Araki, 2012a; Pei et al., 2014). Intriguingly, endogenous or overexpressed Rab20 associates with mitochondria (Hackenbeck et al., 2011).
Here we seek to define the function of Rab20 in the endocytic pathway and its links to cytokine activation. We find that in macrophages, Rab20 associates with early endocytic organelles, and its expression induces the formation of enlarged endosomes. Of importance, both the expression of Rab20 and the enlargement of Rab20-positive endosomes are triggered by IFN-γ activation. By live-cell imaging, we find that Rab20 expression stimulates the homotypic fusion of early endosomes. Finally, whereas both endocytic uptake of transferrin and macropinocytosis are not affected by the expression of the wild-type and dominant-negative forms of Rab20, Rab20 knockdown accelerates epidermal growth factor (EGF)–stimulated endocytic trafficking to LAMP-2–positive compartments and degradation of EGF receptor (EGFR). Taking the results together, we define the function of Rab20 in the endocytic pathway during activation of macrophages with IFN-γ.
RESULTS
Rab20 is associated with the Golgi complex and early endosomes in resting macrophages
Little is known about the localization of endogenous Rab20. We found, using a specific antibody (Supplemental Figure S1), that in resting RAW264.7 macrophages, endogenous Rab20 localized to the cis-Golgi network, trans-Golgi network, early endosomes, and, to a lesser extent, late endosomes or lysosomes (Supplemental Figure S2). At the ultrastructural level, immunogold labeling of ultrathin thawed cryosections confirmed that endogenous Rab20 is associated with the Golgi complex (Figure 1A). To investigate Rab20 localization on endocytic structures, we preloaded cells with 5-nm bovine serum albumin (BSA)–gold for 1 h to fill the endocytic compartments. We found that endogenous Rab20 colocalized with the 5-nm BSA-gold–filled endosomes, including multivesicular body structures (Figure 1B). Further, cells were preloaded with 5-nm BSA-gold for 15 min to label predominantly early endosomes. In these cells, Rab20 labeling was found on the 5-nm BSA-gold–filled early endosomes (Figure 1, C–E). Taken together, these data show that endogenous Rab20 is mainly associated with the Golgi complex and early endosomes in resting macrophages.
FIGURE 1:
Rab20 distribution in macrophages at the ultrastructural level. (A) RAW264.7 macrophages preloaded with 5-nm BSA-gold for 15 min. (B) RAW264.7 macrophages were preloaded with 5-nm BSA-gold for 1 h. (C–E). RAW264.7 macrophages were preloaded with 5-nm BSA-gold for 15 min. Samples were then processed for cryosectioning, and Rab20 was localized using a rabbit anti-Rab20 antibody followed by protein A-gold (10 nm) labeling on Golgi complex (A), endosomes (B), and early endosomes (C–E). (i, ii) Regions of interest indicated by white rectangles. White arrows indicate Rab20 labeling on 5-nm BSA-gold– preloaded organelles. Scale bar, 500 nm.
Rab20 expression induces the enlargement of early and late endosomes
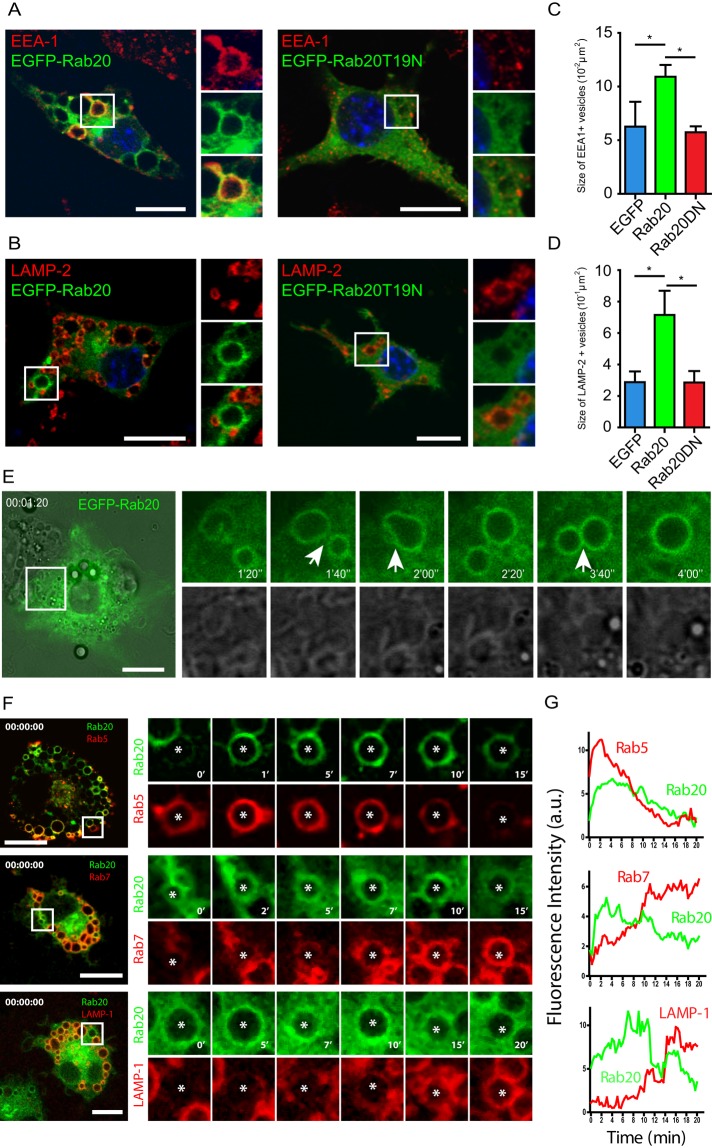
When enhanced green fluorescent protein (EGFP)–Rab20 was overexpressed, we found that, similar to endogenous Rab20, EGFP-Rab20 mainly associated with the cis- and trans-Golgi network and early endosomes but not late endosomes or lysosomes (unpublished data). Strikingly, we observed that expression of EGFP-Rab20 induced vesicle enlargement in macrophages without affecting cell viability (unpublished data). Large vacuoles were positive for both the early endosomal marker EEA-1 and EGFP-Rab20 (Figure 2A). We also observed enlarged vacuoles positive for the late endosomal marker LAMP-2 but negative for EGFP-Rab20 (Figure 2B). Consistently, EGFP-Rab20–positive enlarged vacuoles were not positive for LysoTracker, whereas large vacuoles negative for Rab20 were LysoTracker positive (Supplemental Figure S3). In contrast, overexpression of EGFP or the dominant-negative mutant EGFP-Rab20T19N had no effect on the size of early or late endosomes (Figure 2, A and B). A quantitative analysis of EEA-1– or LAMP-2–positive structures revealed that the average area of early and late endosomes increased approximately twofold in macrophages expressing EGFP-Rab20 (Figure 2, C and D). To investigate the process of the formation of these large EGFP-Rab20–positive vacuoles, we performed live-cell imaging in RAW264.7 macrophages expressing EGFP-Rab20. Very often, we observed homotypic fusions between Rab20-positive endosomes (Figure 2E, white arrows). Of interest, in macrophages coexpressing mCherry-Rab20 and EGFP-Rab5, we found that Rab20 associates to large endosomes after Rab5 and that large endosomes were observed at different stages: Rab5 positive, Rab5/Rab20 positive hybrid endosomes, or Rab20 positive (Figure 2F). Conversely, in cells coexpressing mCherry/EGFP-Rab20 with either EGFP-Rab7 or tdTomato-LAMP-1, Rab20 precluded both Rab7 and LAMP-1 association (Figure 2, F and G). Taken together, the data argue that Rab20 expression induces homotypic fusion of early endosomes, leading to an increase in the size of the endocytic compartment.
FIGURE 2:
EGFP-Rab20 expression induces the enlargement of early and late endosomes in macrophages. Cells were transfected with EGFP-Rab20 or EGFP-Rab20T19N and subsequently immunolabeled for (A) the early endosomal marker EEA-1 or (B) the late endosomal marker LAMP-2. Nuclei were stained with Hoechst 33258 and are shown in blue. Insets show regions of interest indicated by the white rectangles. Scale bar, 10 μm. Quantitative analysis of the size of (C) early and (D) late endosomes in EGFP–, EGFP-Rab20–, or EGFP-Rab20T19N–expressing RAW264.7 macrophages. For each group, at least 20 cells were analyzed. Data show mean ± SD of three independent experiments. The p values were calculated using Student's two-tailed t test. *p ≤ 0.05. (E) Left, snapshot of RAW264.7 macrophages expressing EGFP-Rab20 analyzed by live-cell imaging. Scale bar, 10 μm. Right, the process of homotypic fusions between Rab20-positive endosomes. White arrows indicate fusion events. (F) Left, snapshots at time 0 of RAW264.7 macrophages coexpressing mCherry- or EGFP-Rab20 with EGFP-Rab5, EGFP-Rab7 (pseudocolored in red), and tdTomato-LAMP-1; white squares show zoomed-in areas on the right. Right, zoomed snapshots at the indicated time points of vesicles (asterisks). Scale bar, 10 μm. (G) The kinetics of Rab20 and Rab5, Rab7 or LAMP-1 association with endosomes shown in F.
Rab20 expression and membrane association are induced by IFN-γ activation
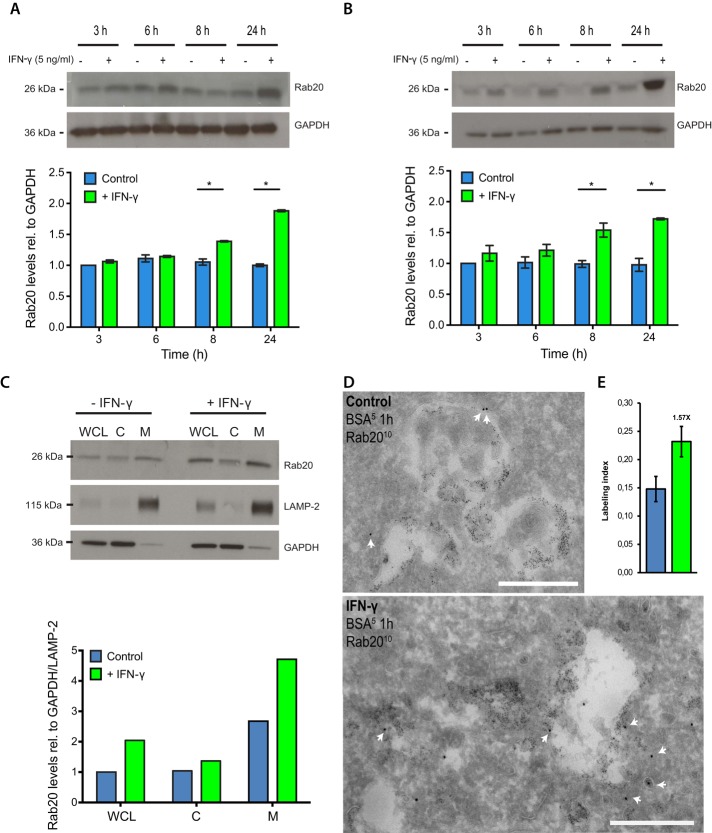
IFN-γ significantly increases the association of Rab20 with phagosomes in macrophages (Pei et al., 2012). To investigate whether Rab20 expression is regulated by IFN-γ, we treated RAW264.7 or primary bone marrow macrophages (BMMs) with or without IFN-γ and analyzed the expression level of Rab20 by Western blotting. In both RAW264.7 macrophages and BMMs, the expression levels of Rab20 increased approximately twofold after 24 h of IFN-γ treatment relative to resting cells (Figure 3, A and B), similar to the levels reached with the expression of EGFP-Rab20 (Supplemental Figure S4). Of importance, this increase in Rab20 levels was reflected in the amount of membrane-bound Rab20 (Figure 3C). Consistent with these observations, we also observed by electron microscopy that the levels of Rab20 immunogold labeling increased, particularly in endosomes. Quantitative analysis using stereology showed that the intensity of Rab20 labeling on BSA-gold–filled endosomes increased ∼1.6-fold after IFN-γ treatment (Figure 3, D and E). Taking the results together, we conclude that IFN-γ induced both the expression and endosomal association of Rab20 in macrophages.
FIGURE 3:
Rab20 expression is increased by IFN-γ treatment. (A) Western blot analysis of Rab20 expression after IFN-γ treatment in RAW264.7 macrophages. (B) Western blot analysis of Rab20 expression after IFN-γ treatment in bone marrow macrophages. RAW264.7 macrophages or bone marrow macrophages were treated with IFN-γ for the indicated times, and cell lysates were collected for Western blotting. Relative intensity was calculated considering the loading control and normalized to the relative intensity in untreated sample at 3 h. Data show mean ± SD of three independent experiments. The p values were calculated using Student's two-tailed t test. *p ≤ 0.05. (C) Western blot analysis of Rab20 levels in whole-cell lysate (WCL), cytosol fraction (C), and membrane fraction (M) with or without IFN-γ treatment in RAW264.7 macrophages. (D) RAW264.7 macrophages were treated without (top) or with (bottom) IFN-γ as described, then preloaded with 5-nm BSA-gold for 1 h and processed for cryosectioning. Rab20 was detected using a rabbit anti-Rab20 antibody, followed by protein A-gold (10 nm) labeling. White arrows indicate Rab20 labeling of endosomes. Scale bar, 500 nm. (E) Stereological analysis of Rab20 labeling intensity on 5-nm BSA-gold–preloaded organelles. Three grids were analyzed for each sample; data show mean ± SD. Results for one representative experiment are shown out of two experiments.
IFN-γ induces the enlargement of early endosomes and late endosomes
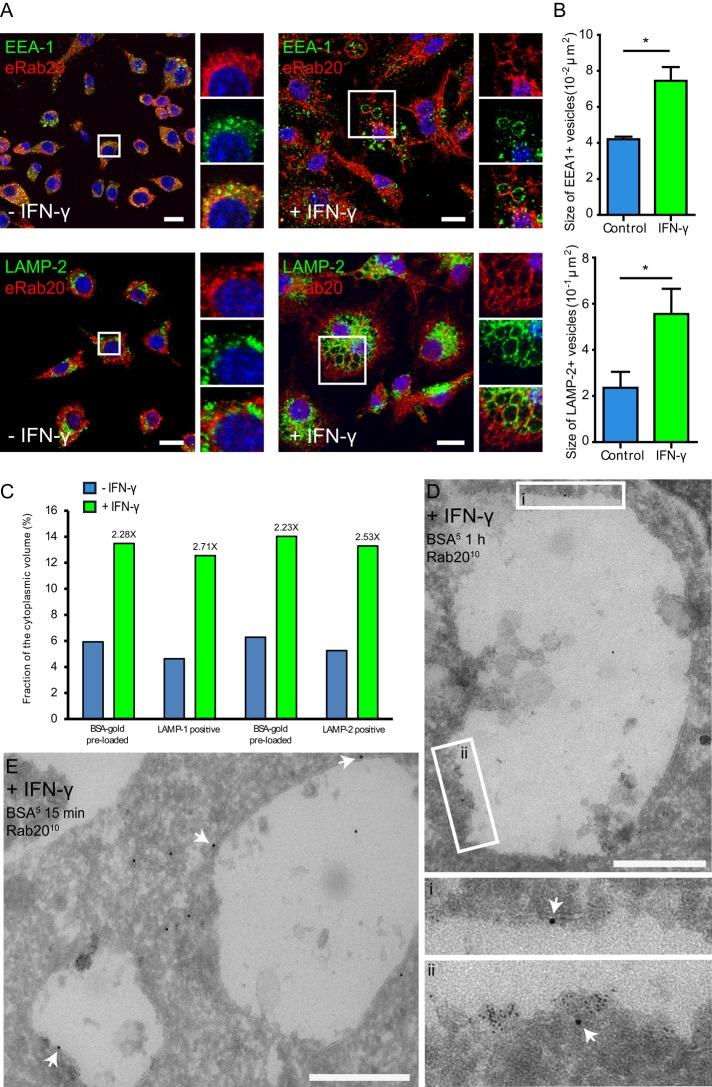
We next investigated the distribution of Rab20 in IFN-γ–activated macrophages. Strikingly, IFN-γ induced an enlargement of the endocytic structures in macrophages. Enlarged early endosomes induced by IFN-γ were double labeled for Rab20 and EEA-1, whereas the enlarged late endosomes induced by IFN-γ were positive for the late endocytic marker LAMP-2 but not Rab20 (Figure 4A). A quantitative analysis indicated that the average area of EEA-1–positive or LAMP-2–positive endosomes was significantly increased by IFN-γ treatment (Figure 4B and Supplemental Figure S5). Further quantification on LAMP-1 or LAMP-2 immunogold-labeled sections of cells preloaded with 5-nm BSA-gold confirmed that the relative volume of LAMP-1– or LAMP-2–positive or BSA-gold–preloaded compartments increased approximately twofold (Figure 4C). Consistent with these observations, electron microscopy immunogold labeling revealed that Rab20 was associated with enlarged endosomes (Figure 4, D and E). Taken together, our data indicate that IFN-γ induces the enlargement of Rab20-positive early endosomes. Moreover, our data also suggest that although Rab20 dissociates from early large endosomes, both early and late endosomes are enlarged.
FIGURE 4:
IFN-γ treatment induces the enlargement of early and late endosomes in macrophages. (A) Distribution of endogenous Rab20 in resting (left) or IFN-γ–activated (right) macrophages. RAW264.7 macrophages were treated without or with 200 U/ml IFN-γ for 16–20 h, fixed, and double stained for Rab20 and the early endosomal marker EEA-1 or the late endosomal marker LAMP-2. Nuclei were stained with Hoechst 33258 and are shown in blue. Insets show regions of interest indicated by the white rectangles. Scale bar, 10 μm. (B) Quantitative analysis of the area of early and late endosomes in resting or IFN-γ–activated RAW264.7 macrophages. Early endosomes and late endosomes represent EEA1-positive and LAMP-2-positive structures, respectively. For each group, at least 20 cells were analyzed. Data show mean ± SD of three independent experiments. The p values were calculated using Student's two-tailed t test. *p ≤ 0.05. (C) Stereological analysis of the relative volume of 5-nm BSA-gold–preloaded, LAMP-1– or LAMP-2xpositive endosomes in resting or IFN-γ–activated macrophages. Estimate for 5-nm BSA-gold–preloaded endosome was made separately on LAMP-1– or in LAMP-2–labeled sections. (D) Rab20 immunogold labeling on endosomes in IFN-γ–activated macrophages. RAW264.7 macrophages were treated with IFN-γ and then preloaded with 5-nm BSA-gold for 15 min to label mostly early endosomes and processed for cryosectioning. Rab20 was detected using a rabbit anti-Rab20 antibody, followed by protein A-gold (10 nm) labeling. White arrows indicate Rab20 labeling on enlarged endosomes. Scale bar, 500 nm. (E) The same as in D, but the cells were preloaded with 5-nm BSA-gold for 1 h to label the entire endocytic pathway. (i, ii) Regions of interest indicated by white rectangles.
Rab20 is required for IFN-γ–dependent enlargement of endosomes
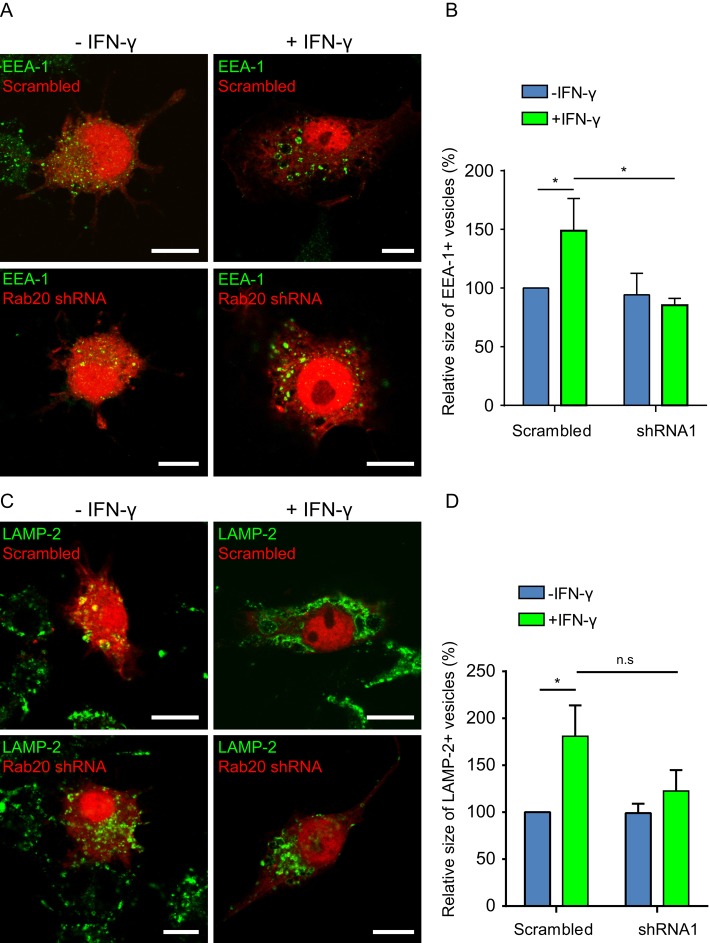
Next we investigated whether the IFN-γ–dependent enlargement of the endocytic compartment requires Rab20. We quantified the relative size of endocytic organelles in both macrophages expressing either scrambled short hairpin RNA (shRNA) or Rab20 shRNA. We observed that in the scrambled control, the EEA-1–positive compartment was significantly larger after IFN-γ treatment (Figure 5, A and B). This effect was significantly abrogated in cells knocked down for Rab20 (Figure 5, A and B). When we analyzed the size of the LAMP-2–positive compartment, we found that IFN-γ induced a significant enlargement of this compartment as well (Figure 5, C and D). Although there was a substantial decrease in the size of the LAMP-2 compartment after IFN-γ activation in Rab20 knockdown cells, it did not reach statistical significance (Figure 5, C and D). Thus the effect of IFN-γ on the regulation of the size of the early endocytic compartment through homotypic fusion requires Rab20.
FIGURE 5:
The IFN-γ–dependent enlargement of the endocytic compartment in macrophages requires Rab20. (A) Distribution of EEA-1 in macrophages expressing scrambled shRNA (top) or Rab20 shRNA (bottom) with or without IFN-γ treatment. RAW264.7 macrophages expressing scrambled shRNA or Rab20 shRNA were treated with or without IFN-γ for 16–20 h and fixed for immunofluorescence. Scale bar, 10 μm. (B) Quantitative analysis of the area of EEA-1–positive structures in macrophages expressing scrambled shRNA or Rab20 shRNA with or without IFN-γ treatment. For each group, at least 20 cells were analyzed. Data show mean ± SD of three independent experiments. The p values were calculated using Student's two-tailed t test. *p ≤ 0.05. (C) Distribution of endogenous LAMP-2 in macrophages expressing scrambled shRNA (top) or Rab20 shRNA (bottom) with or without IFN-γ treatment. RAW264.7 macrophages expressing scrambled shRNA or Rab20 shRNA were stimulated with IFN-γ or left untreated for 16–20 h and fixed for immunofluorescence. Scale bar, 10 μm. (D) Quantitative analysis of the area of LAMP-2–positive structures in macrophages expressing scrambled shRNA or Rab20 shRNA with or without IFN-γ treatment. For each group, at least 20 cells per experiment were analyzed. Data show mean ± SD of three independent experiments. The p values were calculated using Student's two-tailed t test. *p ≤ 0.05.
Rab20 knockdown accelerates endocytic trafficking to LAMP-2– positive compartments
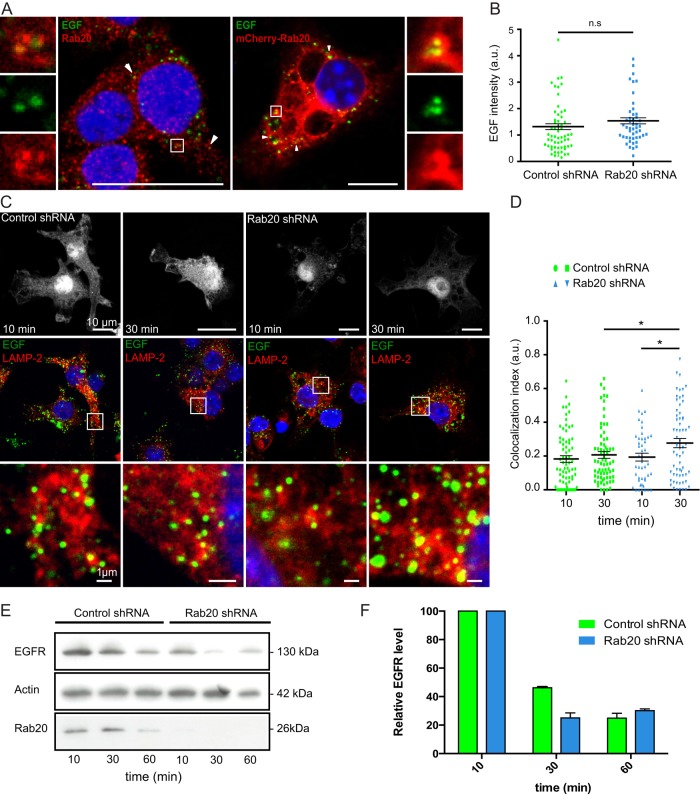
Considering that Rab20 is an endocytic Rab GTPase, we next asked whether Rab20 is functionally linked to this pathway. We found no significant differences in the amount of internalized transferrin and dextran 70 kDa in macrophages expressing EGFP, EGFP-Rab20, or EGFP-Rab20DN (Supplemental Figure S6). Then we investigated the role of this GTPase in endosome trafficking. Alexa Fluor 488–conjugated epidermal growth factor (EGF) was internalized by macrophages expressing either scrambled shRNA or Rab20 shRNA (Pei et al., 2012). Consistent with our previous experiments (Supplemental Figure S6), EGF was localized in Rab20-positive endosomes (Figure 6A), but the internalization of EGF was not affected in Rab20 KD macrophages (Figure 6B). In contrast, Rab20 knockdown significantly increased the colocalization of EGF with LAMP-2–positive compartments compared with macrophages expressing scrambled shRNA (Figure 6, C and D). Furthermore, knockdown of Rab20 accelerated the degradation of EGFR compared with the controls (Figure 6, E and F). Taken together, these results suggest that whereas Rab20 does not participate in endocytic uptake, Rab20 knockdown accelerates EGF-stimulated endosome maturation, lysosomal targeting, and degradation.
FIGURE 6:
Rab20 knockdown accelerates endocytic trafficking and degradation in macrophages. (A) Colocalization of EGF with endogenous Rab20 (left) or overexpressed Rab20 (right) after 10 min of internalization. Insets show regions of interest indicated by the white rectangles. Scale bar, 10 μm. (B) Quantification of internalized EGF at 10 min of incubation. For each group, at least 50 cells were analyzed. Data show mean ± SEM of one representative experiment from three independent experiments; ns: nonsignificant. (C) EGF-stimulated endocytosis and postendocytic trafficking in macrophages expressing scrambled shRNA or Rab20 shRNA. Alexa Fluor 488–conjugated EGF was added to macrophages for 1 h on ice and incubated for the indicated time at 37°C. After fixation, cells were stained for LAMP-2. Insets show regions of interest indicated by the white rectangles. Scale bar, 10 μm. (D) Quantification of the colocalization of Alexa Fluor 488–conjugated EGF and LAMP-2. For each group, at least 50 cells were analyzed. Data show mean ± SEM of one representative experiment from three independent experiments. The p values were calculated using Student's two-tailed t test. *p ≤ 0.05. (E) Western blot analysis of EGFR levels upon EGF stimulation in macrophages expressing scrambled shRNA or Rab20 shRNA. RAW264.7 macrophages stably expressing scrambled shRNA or Rab20 shRNA were incubated with EGF for 1 h on ice and incubated for the indicated time at 37°C. Cells were collected, and cell lysates were blotted with anti-EGFR antibody. Actin was used as loading control. (F) Quantification of EGFR levels in macrophages expressing scrambled shRNA or Rab20 shRNA at different time points. Relative intensity of EGFR was calculated considering the loading control. Data show mean ± SEM of two representative experiments.
DISCUSSION
In this study, we characterized in detail the localization of endogenous Rab20 in resting and activated macrophages. We found that in macrophages, Rab20 was mainly localized in the Golgi complex and early endosomes. Similarly, EGFP-Rab20 was also associated with the same compartments. We show that Rab20 overexpression in macrophages induced a striking enlargement of early and late endosomes. Rab20 is localized to these enlarged early endosomes, supporting the idea that Rab20 may function in endocytic trafficking. Both Rab5 and Rab22a are localized in early endosomes (Chavrier et al., 1990; Kauppi et al., 2002), and expression of the constitutively active mutant Rab5-Q79L or Rab22a-Q64L (Stenmark et al., 1994; Roberts et al., 1999; Mesa et al., 2001; Wegner et al., 2010) and the MHC class II–associated invariant chain (Stang and Bakke, 1997; Nordeng et al., 2002) results in the formation of enlarged early and late endosomal compartments. In vitro experiments and live-cell imaging demonstrated that the homotypic fusion between early endosomes in Rab5-Q79L–expressing cells resulted in the formation of giant endosomes (Stenmark et al., 1994; Roberts et al., 1999). In agreement with these data, by live-cell imaging we also observed the frequent homotypic fusion events between Rab20-positive vesicles in macrophages expressing Rab20. Because Rabex-5 is an effector of Rab20, it is conceivable that Rab20 regulates both phagosome (Pei et al., 2014) and endosome homotypic fusions by increasing the levels of active Rab5 in those organelles. Consistent with this idea, we found that in large endosomes, Rab20 is associated with Rab5-positive endosomes and remains in endosomes after Rab5 dissociation. Moreover, in large endosomes, Rab20 dissociation precluded the recruitment of the late endocytic markers Rab7 and LAMP-1. Supporting this notion, large Rab20-positive endosomes are negative for LysoTracker, whereas large endosomes that were Rab20 negative were LysoTracker positive. We propose that in endosomes, Rab20 recruits active Rab5 as reported in phagosomes (Pei et al., 2014), leading to homotypic fusion and early endosomal enlargement. As a consequence, the late endosomal compartment is also enlarged, as previously reported in cells expressing active Rab5 (Wegner et al., 2010). Future studies are required to define whether this increase in homotypic fusion events is also mediated via Rab20-dependent Rabex-5 recruitment to endosomes. Of interest, in the live-cell imaging studies with Rab5 and Rab20, we often observed Rab20 and Rab5 in the same membrane domains but also in distinct and separate subdomains, suggesting that Rab20 and Rab5 could coordinate functionally different domains and endosomal communication (Sonnichsen et al., 2000).
This work provides unambiguous evidence that Rab20 is an IFN-γ–inducible Rab GTPase in macrophages. However, the regulation of Rab20 expression is rather more complex and involves the transcription factors NF-κB and HIF-1 as well (Gutierrez et al., 2008; Hackenbeck et al., 2011). Collectively these data indicate that Rab20 expression is highly regulated, although it remains to be defined how all of these signals converge to modulate the expression of Rab20. Our data indicate that Rab20 is a critical part of the molecular machinery involved in the previously observed IFN-γ–induced alterations in the formation and morphology of endosomes (Montaner et al., 1999; Tsang et al., 2000). Here we also revealed part of the molecular machinery by which endocytic trafficking is regulated by IFN-γ. In our study, enlarged endosomes were also induced in IFN-γ–activated RAW264.7 macrophages. Some of the enlarged vesicles were positive for the early endosomal marker EEA-1 and others for the late endosomal marker LAMP-2, resembling the phenotype observed in macrophages expressing Rab20. Thus it is likely that increased levels of Rab20 induced by IFN-γ result in enhanced homotypic fusion of early endosomes, which leads to their enlargement. However, an additional role for Rab5 in this process could not be excluded, since its expression is also increased by IFN-γ in macrophages (Alvarez-Dominguez and Stahl, 1998), and Rab5 colocalizes with Rab20 on early endosomes.
Finally, we provide for first time insights into the function of this small GTPase in the endocytic pathway. Rab20 is an early endocytic and phagocytic Rab, which functions more in the maturation of early endocytic and phagocytic organelles than it does in the endocytic uptake process. Other studies identified Rab20 associated with endosomes and macropinosomes, suggesting an additional role in these pathways (Lütcke et al., 1994; Egami and Araki, 2012a). However, in these studies, functional analyses of these pathways were not shown. In our study, using a dominant-negative form of Rab20, we found no differences in endocytic or macropinocytic uptake. This suggests that Rab20 may have a role later during maturation of endocytic organelles. In agreement with this idea, EGF-stimulated endosome maturation is accelerated by Rab20 knockdown. It was reported that macropinosome maturation is slowed by IFN-γ treatment (Tsang et al., 2000). Hence Rab20 may be the link between the reduced macropinosome maturation and IFN-γ activation in macrophages, although more studies are clearly needed to precisely determine the mechanism.
In summary, our data demonstrate that Rab20 is an endocytic Rab GTPase regulated by IFN-γ that leads to enhanced homotypic fusion between early endosomes and targets endosomal cargo to lysosomes for degradation. Therefore this work not only assigns a function for Rab20 in the endocytic pathway, but it also contributes to the understanding of the molecular mechanisms by which IFN-γ treatment modulates the endocytic pathway.
MATERIALS AND METHODS
Cells and reagents
RAW264.7 macrophages were obtained from the American Type Culture Collection (#IB-71) and maintained in complete DMEM, 4.5 g/l glucose with 10% (vol/vol) heat-inactivated fetal calf serum (PAA, Austria), and 2 mM l-glutamine (PAA, Pasching, Austria). BMMs were isolated and maintained as described previously (Kasmapour et al., 2012). Cells were incubated at 37°C/5% CO2 in a humidified incubator. For IFN-γ stimulation, RAW264.7 macrophages and BMMs were stimulated with 5 ng/ml IFN-γ (Peprotech, Hamburg, Germany) for 16–20 h. Recombinant mouse EGF was from BioLegend (San Diego, CA). LysoTracker Red DND-99 was from Invitrogen. The mouse Rab20 gene was amplified by standard PCR from mouse DNA with specific primers and cloned into pEGFP-C1 and pmCherry-C1 vectors (Clontech, Carlsbad, CA). pEGFP-C1-Rab20T19N (Rab20DN) plasmid was generated by site-directed mutagenesis using a QuikChange II XL mutagenesis kit (Agilent Genomics, Santa Clara, CA). RNAi-Ready pSIREN-RetroQ-DsRed-Express vector was obtained from Clontech. EGFP-Rab5 was kindly provided by Philip D. Stahl (Washington University, St. Louis, MO), and EGFP-Rab7 and tdTomato-LAMP1 were kindly provided by Bo Van Deurs (University of Copenhagen, Copenhagen, Denmark) and Tom Carter (Medical Research Council-National Institute for Medical Research, London, United Kingdom), respectively.
Antibodies
The following primary antibodies were used: mouse anti–EEA-1 (610457, BD Transduction Laboratories, San Jose, CA); rat anti–LAMP-1 (1D4B; Developmental Studies Hybridoma Bank [DSHB, Iowa City, IA]); rat anti–LAMP-2 (ABL-93; DSHB); rabbit anti-Rab20 (11616-1-AP; Proteintech, Chicago, IL); rabbit anti-Rab20 (GTX119559; GeneTex, Irvine, CA); mouse anti–β-actin antibody (A5316; Sigma-Aldrich, Steinheim, Germany), rabbit anti-GADPH (Sigma-Aldrich), and rabbit anti-EGFR antibody (1005; Santa Cruz Biotechnology, Dallas, TX). Secondary antibodies conjugated with Alexa Fluor 488, 546, or 633 for indirect immunofluorescence studies were from Molecular Probes (Invitrogen). Secondary antibodies conjugated with horseradish peroxidase were obtained from Jackson ImmunoResearch Laboratories and Promega. Unconjugated rabbit anti-rat used for immunogold labeling was from Rockland.
Macrophage transfection
In the case of Lipofectamine 2000 (Invitrogen), 0.5 × 105 cells/well were seeded in a 24-well plate 1 d before transfection. On the day of transfection, cells were washed twice with prewarmed Ca2+/Mg2+-free Dulbecco's phosphate-buffered saline (PBS; PAA) and incubated with Opti-MEM reduced serum medium (Invitrogen). We used 0.5 μg of plasmidic DNA and 1 μl of Lipofectamine 2000 diluted in Opti-MEM for each well. After 6 h, the Opti-MEM medium was changed with complete DMEM. For JetPEI-Macrophage transfection (Polyplus-transfection, Illkirch, France), 1 × 105 cells/well were seeded in a 24-well plate 1 d before transfection. We used 1 μg of plasmidic DNA and 3 μl of JetPEI-Macrophage for each transfection.
Indirect immunofluorescence
Cells were fixed with 4% (vol/vol) paraformaldehyde (PFA; Electron Microscopy Sciences, Hatfield, PA) in PBS, pH 7.4, for 10 min at room temperature, followed by incubation with precooled methanol (J.T. Baker, Center Valley, PA) for 2 min at −20°C. After washing twice with PBS, cells were incubated with 50 mM glycine in PBS, pH 7.4, for 10 min and then with 1% BSA (Sigma-Aldrich) in PBS for 10 min. The primary and secondary antibodies were diluted in PBS at the indicated dilutions and incubated for 1 h at room temperature. We used 1 μg/ml Hoechst 33258 (Sigma-Aldrich) for nuclear staining. Cells were mounted with DAKO mounting medium (Dako Cytomation, Glostrup, Denmark). The confocal images were acquired using Leica SP5 AOBS microscope (Leica Microsystems, Wetzlar, Germany) fitted with a 63× oil objective. All the images were taken at the same zoom factor (2.5×).
Transferrin uptake assay
RAW264.7 macrophages were seeded on coverslips in 24-well plates 1 d before transfection. The cells were transfected with EGFP, EGFP-Rab20, or EGFP-Rab20DN with Lipofectamine 2000. After 16–20 h, cells were washed twice with PBS and incubated in serum-free DMEM for 1 h at 37°C. The cells were then washed twice with ice-cold PBS and added with ice-cold serum-free DMEM containing 25 μg/ml Alexa Fluor 647–conjugated transferrin (Invitrogen). The cells were immediately transferred to a 37°C incubator and incubated for 15 min. After washing with PBS, the cells were fixed with 3% (vol/vol) PFA for 10 min at room temperature.
Dextran 70 kDa uptake assay
RAW264.7 macrophages were seeded on coverslips in 24-well plates 1 d before transfection. The cells were transfected with EGFP, EGFP-Rab20, or EGFP-Rab20DN with Lipofectamine 2000. After 16–20 h, the cells were washed twice with PBS and incubated in serum-free DMEM containing 100 μg/ml lysine-fixable dextran 70 kDa and Texas red (Invitrogen) for 1 h at 37°C. After washing with PBS, the cells were fixed with 3% (vol/vol) PFA for 10 min at room temperature.
Rab20 knockdown with pSIREN shRNA system
pSIREN plasmids for Rab20 knockdown were constructed as described previously (Pei et al., 2012). For Rab20 knockdown, RAW264.7 macrophages were transfected with pSIREN plasmids containing shRNA specific for mouse Rab20 (5′-CTGACAGAAACAGCCAACA-3′) or scrambled shRNA using JetPEI-Macrophage. After 30 h posttransfection, the cells were used for EGF endocytosis assay.
Epidermal growth factor endocytosis assay
RAW264.7 macrophages were seeded on coverslips in 24-well plates 1 d before transfection. The cells were transfected with scrambled shRNA or shRNA1 against Rab20 using JetPEI-Macrophage. After 30 h, the cells were washed twice with PBS and starved in serum-free DMEM for 5 h at 37°C. The cells were then washed twice with ice-cold PBS and incubated on ice in uptake medium (2% BSA and 20 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid [HEPES], pH 7.5, in serum-free DMEM) containing 5 μg/ml EGF conjugated to Alexa Fluor 488 streptavidin (Invitrogen) for 1 h. After incubation, cells were washed three times with ice-cold PBS to remove unbound ligands. Afterward, the cells were transferred to a 37°C incubator and incubated for the indicated time periods. The cells were fixed and stained with anti–LAMP-2 antibody for confocal microscopy analysis.
Cryosectioning and immunolabeling
RAW264.7 macrophages were fed with 5-nm BSA-gold for 15 min to fill early endosomes or 1 h to fill the entire endocytic pathway, washed with PBS containing 1% BSA, and fixed with 4% PFA in 200 mM HEPES, pH 7.4, overnight. Cell pellet was embedded in 12% bovine gelatin (Sigma-Aldrich, USA) cut into small cubes and infiltrated with 2.3 M sucrose overnight. Samples were vitrified in liquid nitrogen, and 80-nm Tokuyasu sections were cut with a Leica EM UC7 Cryo-ultramicrotome (Leica Microsystems). Sections were transferred on Formvar/carbon-coated transmission electron microscopy (TEM) grids. Cold-water fish skin gelatin (FSG), 1% (Sigma-Aldrich, Germany), in PBS was used for blocking of nonspecific binding and dilution of the bridging antibody and protein A-gold (PAG; Department of Cell Biology, University Medical Center Utrecht, Utrecht, Netherlands). Tris buffer, 10 mM, with 1% FSG, pH 8.0, was used for dilution of Rab20 antibody; it was used at 1:10 dilution. LAMP-1 and LAMP-2 were used undiluted, a bridging rabbit anti-rat antibody was used at 1:250 dilution, and PAG, 10 nm, was used at 1:50 dilution. All labeling reagents were incubated on sections for 30 min. Sections were embedded in 2% methyl cellulose with 0.2% uranyl acetate (Electron Microscopy Sciences) and analyzed with a Philips CM100 TEM microscope (Philips, Amsterdam, Netherlands). The images were recorded digitally with a Quemesa TEM charge-coupled device camera (Olympus Soft Imaging Solutions, Hamburg, Germany) and iTEM software (Olympus Soft Imaging Solutions).
Stereology
To estimate the labeling index of Rab20, systematic uniform random sampling was carried out in images at 15,500× magnification. Three grids were analyzed per sample. At least 20 images/grid were quantified. The labeling index was calculated as the number of the gold particles on the gold-filled compartment per area over that compartment as determined by test point counting (Lucocq et al., 2004). To estimate the relative volume of BSA-gold–preloaded LAMP-1– or LAMP-2xpositive organelles, at least 20 images were collected by systematic uniform random sampling at 11,500× magnification. By test point counting, the relative volume was estimated as the fraction of the area over these compartments in the area of the cytoplasm.
SDS–PAGE and Western blotting
Cells were lysed in lysis buffer (20 mM Tris, pH 7.5, 150 mM NaCl, 0.5 mM EDTA, 0.5% NP-40) supplemented with complete protease inhibitor cocktail (Roche, Basel, Switzerland) on ice for 10 min. After centrifugation at 16,000 × g for 10 min, the supernatants were heated with SDS sample buffer at 96°C for 10 min. Proteins were separated on a 10% SDS gel and transferred onto nitrocellulose membranes. The membranes were incubated with primary and secondary antibodies in 5% skimmed milk (Carl Roth, Karlsruhe, Germany) diluted in PBS/0.1% (vol/vol) Tween-20 for 1 h at room temperature, respectively. For Rab20 detection, the membranes were incubated with Rab20 antibody (Proteintech) in 5% skimmed milk in PBS/0.1% (vol/vol) Tween-20 overnight at 4°C. The membranes were developed with ECL Detection Kit (GE Life Sciences, Pittsburgh, PA), scanned, and evaluated using ImageJ software (National Institutes of Health, Bethesda, MD). The antibody against β-actin was used as the loading control.
Membrane fractionation
RAW264.7 macrophages were stimulated with 5 ng/ml mouse IFN-γ for 24 h. Cells were harvested (800 × g, 5 min) and homogenized in homogenization buffer (250 mM sucrose, 0.5 M ethylene glycol tetraacetic acid, 20 mM HEPES, KOH, pH 7.2) by passing 20× through a 27-gauge needle connected to a syringe. The postnuclear fraction was obtained by centrifugation at 1200 × g (10 min, 4°C). Membrane fractions were separated from cytosol by ultracentrifugation of the postnuclear fraction at 100,000 × g for 1 h at 4°C (Beckmann TLA-110 Rotor, 50,000 rpm), followed by two washing steps with 1 M NaCl. Twenty-microgram protein samples were run on a 4–20% Tris-glycine polyacrylamide gel (Invitrogen).
EGFR degradation assay
Rab20-knockdown RAW264.7 macrophages were produced using a MISSION shRNA system (Sigma-Aldrich, USA) as described previously (Pei et al., 2012). Cells stably transfected with Rab20 shRNA oligo 2643 (5′-CCTTTACAAGAAGATCCTGA-3′) or empty control shRNA plasmid pLKO.1 were seeded 2 d before experiment in T25 flasks. Then cells were washed twice with PBS and starved in serum-free DMEM for 5 h at 37°C. For EGF uptake, cells were washed twice with ice-cold PBS and incubated for 1 h on ice in uptake medium (2% BSA and 20 mM HEPES, pH 7.5, in serum-free DMEM) containing 2.5 μg/ml recombinant EGF (BioLegend). After incubation, cells were washed three times with ice-cold PBS to remove unbound ligands. PBS was replaced by uptake medium, and flasks were transferred to a 37°C incubator and incubated for the indicated time periods (10, 30, and 60 min). Then cells were washed with PBS, scraped, and processed for Western blot.
Image analysis
To quantify the fluorescence intensity of transferrin or dextran 70 kDa, the overlay images were loaded into ImageJ, version 1.43u, and the different channels were separated. The EGFP–, EGFP-Rab20–, or EGFP-Rab20DN–expressing cells were selected with the Polygon selections tool in the green channel. The fluorescence intensity of transferrin or dextran 70 kDa was measured by redirecting to the red channel. To quantify the colocalization index of EGF and LAMP-2, “Just another colocalization plugin” was used (Bolte and Cordelieres, 2006). The images were adjusted with the same settings of threshold. The colocalization index was defined as the fraction of EGF overlapping with LAMP-2 and was calculated with M1 and M2 Manders coefficients. To quantify the size of early and late endosomes/lysosomes, the overlay images were loaded into ImageJ, version 1.43u, and scales were set. Cells for quantification were selected by the Polygon selections tool. Then different channels were separated, and channel EEA-1 or LAMP-2 was adjusted with the same threshold. In Set Measurements, Area and Limit to threshold were selected. Finally, the adjusted images were measured with Analyze particles. The results were exported, and the average size was calculated.
Statistical analysis
Statistical analysis was performed with GraphPad Prism, version 5.04 (GraphPad Software). The p values were calculated using Student's two-tailed t test. The confidence interval was 95%.
Supplementary Material
Acknowledgments
We thank Norbert Roos and the EM Core Facility in the Department of Biosciences, University of Oslo, Oslo, Norway, for their support. This work was funded by the Initiative and Networking funds of the Helmholtz Association, the Medical Research Council UK (MC_UP_1202/11) to M.G.G., and the German Research Council (DFG) SPP1580 to G.G. and M.G.G.
Abbreviations used:
- BMMs
bone marrow macrophages
- BSA
bovine serum albumin
- EGF
epidermal growth factor
- EGFP
enhanced green fluorescent protein
- EGFR
epidermal growth factor receptor
- shRNA
short hairpin RNA.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E14-11-1547) on July 8, 2015.
*These authors contributed equally to this work.
The authors have no conflicting financial interests.
REFERENCES
- Alvarez-Dominguez C, Stahl PD. Interferon-gamma selectively induces Rab5a synthesis and processing in mononuclear cells. J Biol Chem. 1998;273:33901–33904. doi: 10.1074/jbc.273.51.33901. [DOI] [PubMed] [Google Scholar]
- Amillet J-M, Ferbus D, Real FX, Antony C, Muleris M, Gress TM, Goubin G. Characterization of human Rab20 overexpressed in exocrine pancreatic carcinoma. Hum Pathol. 2006;37:256–263. doi: 10.1016/j.humpath.2005.10.017. [DOI] [PubMed] [Google Scholar]
- Barry AO, Mege JL, Ghigo E. Hijacked phagosomes and leukocyte activation: an intimate relationship. J Leukocyte Biol. 2011;89:373–382. doi: 10.1189/jlb.0510270. [DOI] [PubMed] [Google Scholar]
- Bolte S, Cordelieres FP. A guided tour into subcellular colocalization analysis in light microscopy. J Microsc. 2006;224:213–232. doi: 10.1111/j.1365-2818.2006.01706.x. [DOI] [PubMed] [Google Scholar]
- Casbon AJ, Long ME, Dunn KW, Allen LA, Dinauer MC. Effects of IFN-gamma on intracellular trafficking and activity of macrophage NADPH oxidase flavocytochrome b558. J Leukocyte Biol. 2012;92:869–882. doi: 10.1189/jlb.0512244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavrier P, Parton RG, Hauri HP, Simons K, Zerial M. Localization of low molecular weight GTP binding proteins to exocytic and endocytic compartments. Cell. 1990;62:317–329. doi: 10.1016/0092-8674(90)90369-p. [DOI] [PubMed] [Google Scholar]
- Curtis LM, Gluck S. Distribution of Rab GTPases in mouse kidney and comparison with vacuolar H+-ATPase. Nephron Physiol. 2005;100:31–42. doi: 10.1159/000085114. [DOI] [PubMed] [Google Scholar]
- Das Sarma J, Kaplan BE, Willemsen D, Koval M. Identification of rab20 as a potential regulator of connexin 43 trafficking. Cell Commun Adhes. 2008;15:65–74. doi: 10.1080/15419060802014305. [DOI] [PubMed] [Google Scholar]
- Egami Y, Araki N. Rab20 regulates phagosome maturation in RAW264 macrophages during Fc gamma receptor-mediated phagocytosis. PLoS One. 2012a;7:e35663. doi: 10.1371/journal.pone.0035663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egami Y, Araki N. Spatiotemporal localization of Rab20 in live RAW264 macrophages during macropinocytosis. Acta Histochem Cytochem. 2012b;45:317–323. doi: 10.1267/ahc.12014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez MG, Mishra BB, Jordao L, Elliott E, Anes E, Griffiths G. NF-kappa B activation controls phagolysosome fusion-mediated killing of mycobacteria by macrophages. J Immunol. 2008;181:2651–2663. doi: 10.4049/jimmunol.181.4.2651. [DOI] [PubMed] [Google Scholar]
- Hackenbeck T, Huber R, Schietke R, Knaup KX, Monti J, Wu X, Klanke B, Frey B, Gaipl U, Wullich B, et al. The GTPase RAB20 is a HIF target with mitochondrial localization mediating apoptosis in hypoxia. Biochim Biophys Acta. 2011;1813:1–13. doi: 10.1016/j.bbamcr.2010.10.019. [DOI] [PubMed] [Google Scholar]
- Kasmapour B, Gronow A, Bleck CK, Hong W, Gutierrez MG. Size-dependent mechanism of cargo sorting during lysosome-phagosome fusion is controlled by Rab34. Proc Natl Acad Sci USA. 2012;109:20485–20490. doi: 10.1073/pnas.1206811109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauppi M, Simonsen A, Bremnes B, Vieira A, Callaghan J, Stenmark H, Olkkonen VM. The small GTPase Rab22 interacts with EEA1 and controls endosomal membrane trafficking. J Cell Sci. 2002;115:899–911. doi: 10.1242/jcs.115.5.899. [DOI] [PubMed] [Google Scholar]
- Lucocq JM, Habermann A, Watt S, Backer JM, Mayhew TM, Griffiths G. A rapid method for assessing the distribution of gold labeling on thin sections. J Histochem Cytochem. 2004;52:991–1000. doi: 10.1369/jhc.3A6178.2004. [DOI] [PubMed] [Google Scholar]
- Lütcke A, Parton RG, Murphy C, Olkkonen VM, Dupree P, Valencia A, Simons K, Zerial M. Cloning and subcellular localization of novel rab proteins reveals polarized and cell type-specific expression. J Cell Sci. 1994;107:3437–3448. doi: 10.1242/jcs.107.12.3437. [DOI] [PubMed] [Google Scholar]
- Mesa R, Salomon C, Roggero M, Stahl PD, Mayorga LS. Rab22a affects the morphology and function of the endocytic pathway. J Cell Sci. 2001;114:4041–4049. doi: 10.1242/jcs.114.22.4041. [DOI] [PubMed] [Google Scholar]
- Montaner LJ, da Silva RP, Sun J, Sutterwala S, Hollinshead M, Vaux D, Gordon S. Type 1 and type 2 cytokine regulation of macrophage endocytosis: differential activation by IL-4/IL-13 as opposed to IFN-gamma or IL-10. J Immunol. 1999;162:4606–4613. [PubMed] [Google Scholar]
- Nordeng TW, Gregers TF, Kongsvik TL, Meresse S, Gorvel JP, Jourdan F, Motta A, Bakke O. The cytoplasmic tail of invariant chain regulates endosome fusion and morphology. Mol Biol Cell. 2002;13:1846–1856. doi: 10.1091/mbc.01-10-0478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei G, Bronietzki M, Gutierrez MG. Immune regulation of Rab proteins expression and intracellular transport. J Leukocyte Biol. 2012;92:41–50. doi: 10.1189/jlb.0212076. [DOI] [PubMed] [Google Scholar]
- Pei G, Repnik U, Griffiths G, Gutierrez MG. Identification of an immune-regulated phagosomal Rab cascade in macrophages. J Cell Sci. 2014;127:2071–2082. doi: 10.1242/jcs.144923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts RL, Barbieri MA, Pryse KM, Chua M, Morisaki JH, Stahl PD. Endosome fusion in living cells overexpressing GFP-rab5. J Cell Sci. 1999;112:3667–3675. doi: 10.1242/jcs.112.21.3667. [DOI] [PubMed] [Google Scholar]
- Schroder K, Hertzog PJ, Ravasi T, Hume DA. Interferon-gamma: an overview of signals, mechanisms and functions. J Leukocyte Biol. 2004;75:163–189. doi: 10.1189/jlb.0603252. [DOI] [PubMed] [Google Scholar]
- Seto S, Tsujimura K, Koide Y. Rab GTPases regulating phagosome maturation are differentially recruited to mycobacterial phagosomes. Traffic. 2011;12:407–420. doi: 10.1111/j.1600-0854.2011.01165.x. [DOI] [PubMed] [Google Scholar]
- Sonnichsen B, De Renzis S, Nielsen E, Rietdorf J, Zerial M. Distinct membrane domains on endosomes in the recycling pathway visualized by multicolor imaging of Rab4, Rab5, and Rab11. J Cell Biol. 2000;149:901–914. doi: 10.1083/jcb.149.4.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stang E, Bakke O. MHC class II-associated invariant chain-induced enlarged endosomal structures: a morphological study. Exp Cell Res. 1997;235:79–92. doi: 10.1006/excr.1997.3617. [DOI] [PubMed] [Google Scholar]
- Stenmark H. Rab GTPases as coordinators of vesicle traffic. Nat Rev Mol Cell Biol. 2009;10:513–525. doi: 10.1038/nrm2728. [DOI] [PubMed] [Google Scholar]
- Stenmark H, Olkkonen VM. The Rab GTPase family. Genome Biol. 2001;2:REVIEWS3007. doi: 10.1186/gb-2001-2-5-reviews3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenmark H, Parton RG, Steele-Mortimer O, Lütcke A, Gruenberg J, Zerial M. Inhibition of rab5 GTPase activity stimulates membrane fusion in endocytosis. EMBO J. 1994;13:1287–1296. doi: 10.1002/j.1460-2075.1994.tb06381.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torri A, Beretta O, Ranghetti A, Granucci F, Ricciardi-Castagnoli P, Foti M. Gene expression profiles identify inflammatory signatures in dendritic cells. PLoS One. 2010;5:e9404. doi: 10.1371/journal.pone.0009404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang AW, Oestergaard K, Myers JT, Swanson JA. Altered membrane trafficking in activated bone marrow-derived macrophages. J Leukoc Biol. 2000;68:487–494. [PubMed] [Google Scholar]
- Wegner CS, Malerod L, Pedersen NM, Progida C, Bakke O, Stenmark H, Brech A. Ultrastructural characterization of giant endosomes induced by GTPase-deficient Rab5. Histochem Cell Biol. 2010;133:41–55. doi: 10.1007/s00418-009-0643-8. [DOI] [PubMed] [Google Scholar]
- Zerial M, McBride H. Rab proteins as membrane organizers. Nat Rev Mol Cell Biol. 2001;2:107–117. doi: 10.1038/35052055. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.