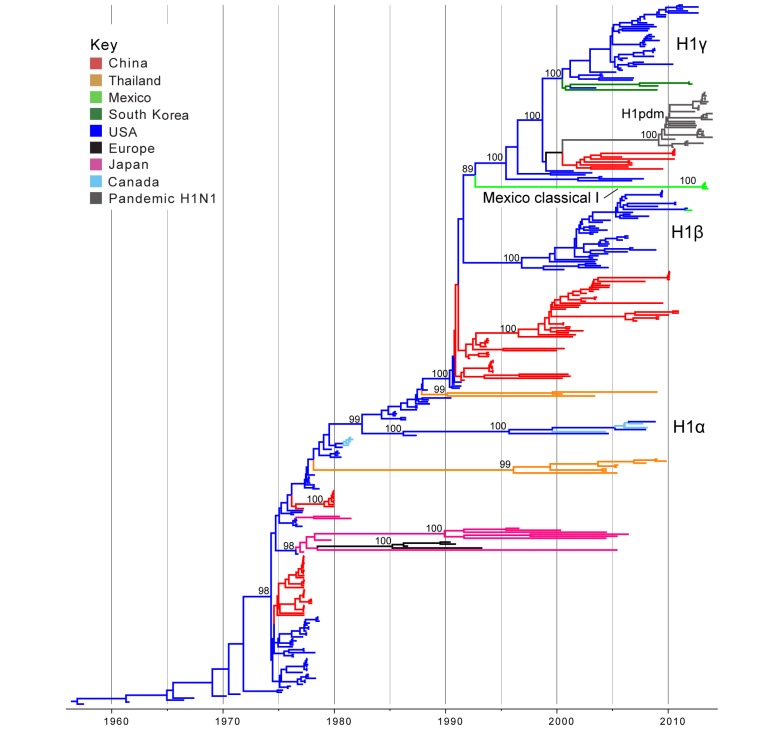
Phylogenetic relationships between classical swine H1 segments.
Time-scaled Bayesian MCC tree inferred for the HA (H1) sequences of 357 viruses, including 5 viruses sequenced for this study from swine in Mexico, 321 classical swine H1 viruses collected globally, and 31 pandemic viruses included as reference. Branches are shaded by country of origin: China = red; Thailand = orange; Mexico = light green; South Korea = dark green; USA/Canada = dark blue; Europe = black; and Japan = pink. As reference, pandemic H1N1 viruses are shaded dark grey. Posterior probabilities > 0.8 are included for key nodes. North American classical IAV-S lineages are labeled: H1α, H1β, and H1γ. The clade of Mexico classical I viruses is labeled (Table 1), and the clade of representative pandemic viruses is labeled ‘H1pdm’ (a detailed phylogeny of H1pdm evolution is available in Fig. S5). The tMRCAs representing the estimated timing of viral introduction into Mexico are provided in parentheses. A similar phylogeny inferred using ML methods with tip labels displayed is available in Fig. S1.