Fig. 1.
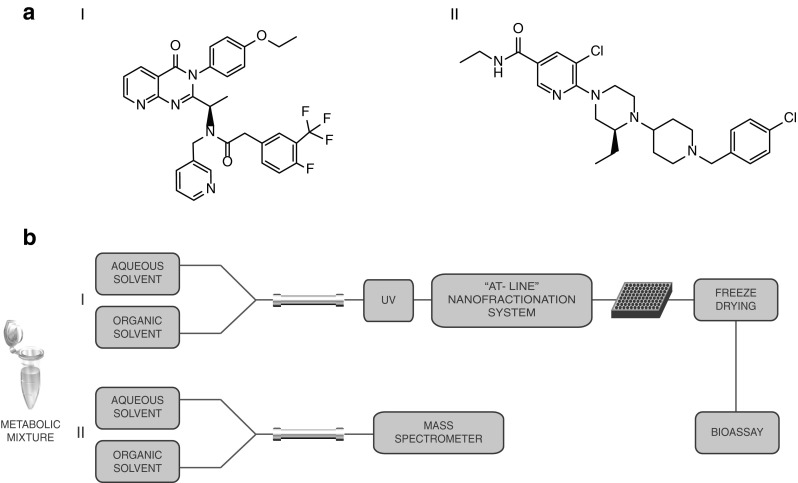
a Chemical structures of two CXCR3 antagonists: I NBI-74330 from the (aza)quinazolinone class II VUF11211 from the piperazinyl-piperidine class. b Schematic overview of the analytical setup. I Separation and UV detection of the metabolic mixtures is followed by high-resolution nanofractionation for bioaffinity screening in radioligand binding assay. II Separation on LC-UV system is followed by accurate MS and MS2 measurements for (partial) structure identification of the metabolites present in the mixture
