The transcription factor NF-κB comprises a family of transcription factors critical for inflammatory signaling, and innate and adaptive immunity.1 NF-κB regulates the transcription of genes involved in immuno-inflammatory responses, cell survival, cell cycle progression, and cell adhesion. Activation of NF-κB by specific ligand-receptor interactions induces the expression of pro-inflammatory mediators, which orchestrate the recruitment of immune cells, induce the inflammatory response, and cause tissue injury.2,3 In this way, excessive NF-κB activity is considered harmful and a pathogenic factor in the chronic inflammation of numerous autoimmune disorders, such as rheumatoid arthritis (RA), systemic lupus erythematosus (SLE), type 1 diabetes, multiple sclerosis, and inflammatory bowel disease.4 Association between overactive NF-κB and autoimmunity emphasizes the importance of tightly controlled NF-κB signaling. One protein that regulates NF-κB signaling and maintains immune homeostasis is the A20 binding inhibitor of NF-κB1 (ABIN1).
ABIN1 is a polyubiquitin binding protein that functions as an inhibitor of NF-κB, JNK, ERK, and p38 MAPK pro-inflammatory signaling pathways.5,6 ABIN1 shares homologous regions with its familial proteins, ABIN2 and ABIN3, that mediate A20-binding and polyubiquitin binding.5,7,8 While the functional role of A20 and polyubiquitin binding is unclear, both have been proposed to participate in ABIN1’s inhibitory mechanism. Disruption of ABIN1’s polyubiquitin binding promotes NF-κB and pro-inflammatory signaling; enhancing the production of pro-inflammatory mediators.5,7 In vivo, this disruption is associated with glomerulonephritis and lupus-like inflammatory disease states.5,9 Currently, the role of ABIN1 in the prevention of human autoimmune and inflammatory disorders is being explored. The focus of this review is to discuss the molecular mechanisms by which ABIN1 functions, and describe its importance for immune homeostasis based on findings from human genetic and animal studies.
ABIN1 expression and structure
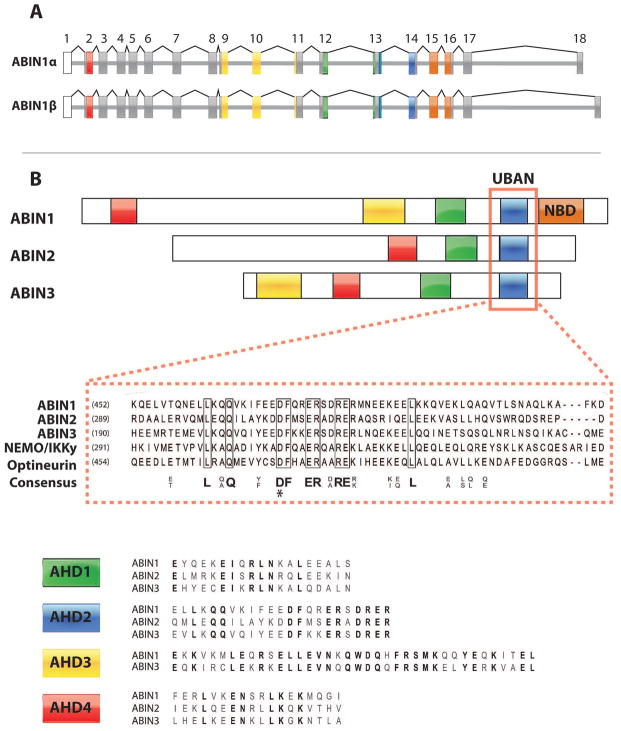
The human gene encoding ABIN1, TNFAIP3 interacting protein 1 (TNIP1), is located on chromosome 5q32-33.1.10 It is comprised of 18 exons, where exons 2–18 contain the coding sequence and exon 1 remains untranslated. Expression of TNIP1 yields two 72 kDa isoforms (ABIN1α and β) which differ in their C termini and are produced by alternative splice acceptor sites within exon 18 (Figure 1A). Moreover, 8 additional alternative splice variants have since been identified.11,12 At the mRNA level, TNIP1 is expressed rather ubiquitously in human tissues with strong expression reported in skeletal muscle, peripheral blood lymphocytes, and spleen.10 TNIP1 mRNA was also found in various hematopoietic immune cell lines, MOLT-4, Jurkat, and HL60. This demonstrates that TNIP1 is well expressed within cells and tissues of the immune system. These findings are intriguing in that TNIP1 polymorphisms have been associated with the development of autoimmune and inflammatory disorders, as described in a later section.
Figure 1. Gene and Domain Structure of ABINs.
A) Splice variation of TNIP1 yields ABIN1α and ABIN1β isoforms, as a result of alternative splice sites within exon 18. The corresponding colors between the exons in figure A and domains in figure B represent exons that encode known ABIN domains: The ABIN Homology Domains (AHDs) 1–4 and NEMO-Binding Domain (NBD). Exons depicted in grey are translated, but do not encode any known domains. Exons depicted in white remain untranslated. B) Sequence alignment between ABIN1 domains and domains of homologous proteins. The UBD in ABIN proteins and NEMO (UBAN) domain is responsible for ABIN1’s ubiquitin binding function and shares sequence homology with NEMO and the NEMO-like protein, Optineurin. AHD1 and AHD2 are required for A20 binding and NF-κB inhibition, respectively. The roles of AHD3 and AHD4 are not described. Within the aligned sequences, bolded letters represent identical, conserved amino acids.
*Mutagenesis (D472N) reported to disrupt ABIN1’s interaction with polyubiquitinated proteins.
Functional domains in ABIN1 have been identified and characterized (Figure 1B). ABIN1 shares regions of high homology with its familial proteins, ABIN2 and ABIN3, termed ABIN Homology Domains (AHD) 1–4. Notably, the AHD2 region of ABIN proteins also share sequence homology with the ubiquitin binding domain (UBD) in NEMO and the NEMO-like protein, optineurin.7 Thus, AHD2 has been renamed the UBD in ABIN proteins and NEMO (UBAN) domain.8 While the current functions of AHD3 and AHD4 are not described, those of AHD1 and UBAN have been explored. Analysis of ABIN1 mutation constructs revealed that AHD1 is responsible for A20 binding whereas the UBAN domain binds to polyubiquitin and polyubiquitinated proteins.5,7,8,13 In addition to AHDs and the UBAN domain, ABIN1 possesses a C-terminal NEMO binding domain (NBD).14 Collectively the UBAN domain, AHD1, and NBD have been shown to participate in the NF-κB inhibitory function of ABIN1.5,8,14
Role of polyubiquitin binding in the inhibition of NF-κB
The UBAN domain of ABIN1 is essential for ubiquitin binding and NF-κB inhibition.5,8,13 Initially by performing yeast two-hybrid screening with ubiquitin as bait, ABINs were identified as novel ubiquitin interacting proteins.8 The affinity of these interactions is dependent on the length of the ubiquitin chains. ABIN1 does not interact with monomeric ubiquitin, but rather with polyubiquitin chains consisting of at least 3 ubiquitin moieties.8,13 Moreover, these UBAN-mediated polyubiquitin interactions are linkage dependent since ABIN1 was shown to bind K63-linked and linear polyubiquitin chains, but not K48-linked polyubiquitin chains.5 This is significant since interactions with K63 and linear moieties mediate signaling events and K48 conjugation mediates protein degradation. This provides the groundwork for the hypothesis that ABIN1’s binding to linkage specific polyubiquitin chains may affect the activity of NF-κB by altering the signal within the pathway. Indeed, ABIN1 interacts with polyubiquitinated, NF-κB mediator proteins NEMO, IRAK1, and RIP1, among other proteins5,8,13 (Table 1) Mutagenesis of conserved amino acids within the UBAN domain (DF485NA, QQ477EE, D472N) diminishes ABIN1’s interaction with these polyubiquitinated proteins.5,8,13 Correspondingly, ABIN1 expressing UBAN mutations fail to abrogate TNF induced NF-κB activation.7 Collectively, this indicates that ubiquitin binding and binding to polyubiquitinated proteins is important for the NF-κB inhibitory function of ABIN1.
Table 1.
ABIN1 interacting proteins.
| Interacting Protein | Interacting Region | Function of Interaction | References |
|---|---|---|---|
| A20 | AHD1 | Inhibition of NF-κB | 6,7 |
| NEMO | NBD | Inhibition of NF-κB | 14 |
| RIP1 | UBAN | Regulates DISC formation in the prevention of apoptosis | 13 |
| TRAF1 | UBAN | Inhibition of TLR-MyD88 signaling | 5 |
| FADD | UBAN | Regulates DISC formation in the prevention of apoptosis | 13 |
| p105 | AHD2 | Inhibition of NF-κB | 46 |
| Nef | HIV infection regulation | 10 | |
| ERK2 | Inhibition of EGF/ERK2 nuclear signaling | 47 |
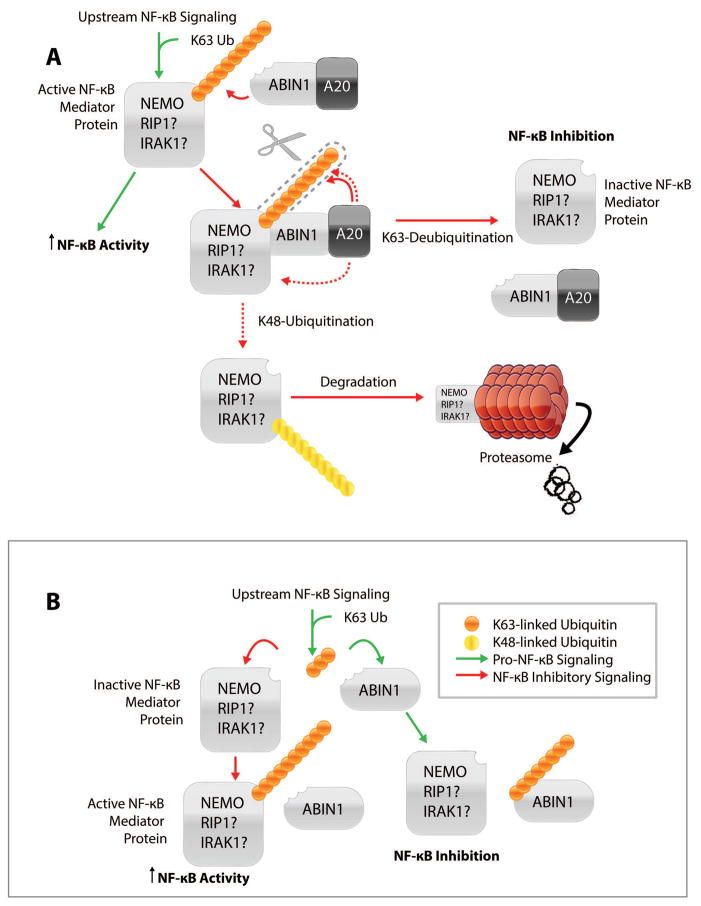
The role of polyubiquitin binding in the NF-κB inhibitory function of ABIN1 is not completely resolved; however, an A20 dependent mechanism is well supported (Figure 2A). ABIN1 is thought to act as an adaptor protein that physically links A20 to polyubiquitinated NF-κB mediators. In this way, ABIN1 is thought to facilitate the activity of A20 as a hydrolase and E3 ligase; cleaving K63-linked and building K48-linked polyubiquitin chains on NF-κB mediators. Therefore, NF-κB activity is inhibited by preventing critical upstream interactions or facilitating proteasome degradation of upstream regulators. This model was, first, proposed on the observation that cells overexpressed with either ABIN1 or A20 display a similar NF-κB inhibitory profile in response to various NF-κB inducers.6 ABIN1, A20, and NEMO were, later, demonstrated to interact as a complex;14 supporting the hypothesis that ABIN1 may cooperatively bind with A20 and NEMO to alter the ubiquitination status of NEMO. Recently, it was shown that while ABIN1 alone could not promote the de-ubiquitination or degradation of NEMO, administering increasing concentrations of A20 promoted NEMO degradation in a dose-dependent manner.15 Likewise, ABIN1 was shown to increase the effects of A20 on NEMO de-ubiquitination.14 Knocking-down ABIN1 with siRNA abrogates A20-dependent de-ubiquitination and results in enhanced NF-κB activity. These findings imply that ABIN1 physically links A20 to NEMO, thereby facilitating the A20-mediated de-ubiquitination of NEMO and inhibition of NF-κB. It should be noted that ABIN1 also interacts with early NF-κB mediators RIP1,and IRAK1.5,13 A20 has also been shown to interact with IRAK1 and remove K63-linked polyubiquitin from NF-κB mediators RIP1 and TRAF6.16–18 Thus, it is likely that ABIN1 may regulate similar A20-mediated, NF-κB inhibitory mechanisms upstream of NEMO.
Figure 2. Mechanisms of ABIN1-Mediated NF-κB Inhibition.
A) A20 dependent mechanism of ABIN-mediated NF-κB inhibition. In this model, ABIN1 acts as an adaptor protein; linking A20 to an active, polyubiquitinated NF-κB mediator protein, such as NEMO. This mechanism of action has been confirmed for NEMO, but simply proposed for NF-κB mediator proteins: RIP1 and IRAK1. Therefore, a question mark follows NF-κB mediator proteins, RIP1 and IRAK1. In this way, ABIN1 facilitates A20’s function as a hydrolase and E3 ligase by mediating the cleavage of K63-linked polyubiquitin and building K48-linked polyubiquitin chains on NF-κB mediator proteins. NF-κB mediator proteins that have had K63-linked ubiquitin cleaved become inactive; resulting in NF-κB inhibition. NF-κB mediator proteins that have had K63-linked ubiquitin cleaved and K48-linked ubiquitin added (dashed red lines) are targeted for degradation in the proteasome; inhibiting NF-κB. B) A20 independent/competitive mechanism of ABIN1-mediated NF-κB inhibition. In this model, ABIN1 competes with NF-κB mediator proteins for polyubiquitin binding. This prevents the ubiquitination and activation of the mediator proteins; inhibiting further NF-κB signaling.
ABIN1 has also been proposed to function independently of A20. ABIN1 deletions lacking AHD1 no longer interact with A20, yet still inhibit NF-κB activity.7 UBAN mutants also demonstrate a dominant-negative effect on the NF-κB inhibitory function of wild type ABIN1, but not of A20. These findings suggest that ABIN1 and A20 can act independently to inhibit NF-κB. A possible A20-independent, NF-κB inhibitory mechanism may be that ABIN1 competes with NF-κB mediator proteins for polyubiquitin (Figure 2B). Indeed, ABIN2 was shown to compete with TRAF6 for K63-linked ubiquitin chains.15 Therefore, through competition ABIN1 may prevent the ubiquitination and activation of NF-κB mediators; preventing further activation of NF-κB. It appears that ABIN1 and A20 may cooperate in some functions but not others. More than likely the mechanism by which ABIN1 acts is cell type, tissue, and/or signal specific.
TNIP1 as a disease susceptibility gene
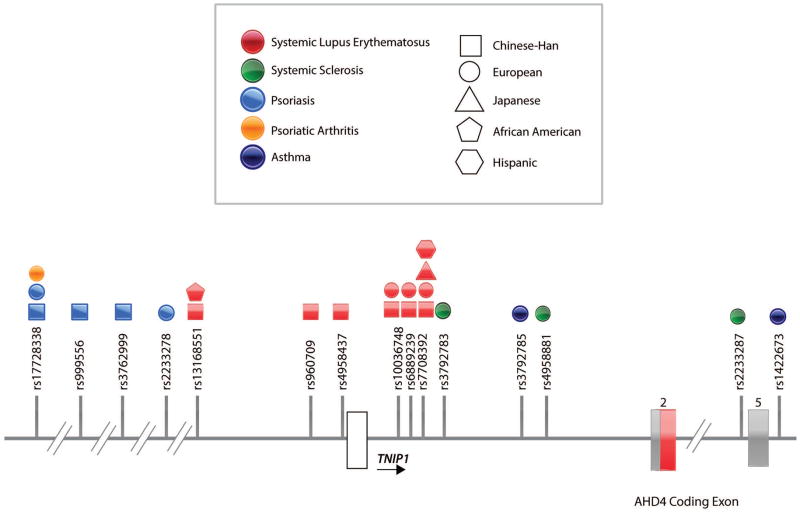
Over the past decade genome wide association studies (GWAS) identified numerous NF-κB susceptibility loci to be associated with autoimmune disease. Among these loci, TNIP1 polymorphisms have been strongly associated with psoriasis, psoriatic arthritis, systemic lupus erythematosus (SLE), and systemic sclerosis (SSc) (Figure 3)19–28,28–31 Nair et al. identified rs17728338 as a risk variant for psoriasis in patients of European ancestry.29 This variant, also, displayed association with psoriatic arthritis; implying a shared genetic etiology between psoriasis and psoriatic arthritis may exist.30 Sun et al. also reported TNIP1 as a susceptibility locus for psoriasis when they identified two variants, rs999556 and rs3762999, to be associated with psoriasis in the Chinese Han population.32 These two variants were found to be in strong linkage disequilibrium (LD) with each other (r2=0.97) and located in the same region as rs17728338.29,32 This suggested that all three variants may link to the same disease association in the Chinese Han population, as well as the European population. By multivariate logistic regression analysis, Sun et al. showed that the associations of rs999556 and rs3762999 were dependent upon the association of rs17728338 in the Chinese Han population.32 However, while they confirmed that rs17728338 is strongly associated with psoriasis in the European population, rs999556 and rs3762999 were not. Thus, the LD among rs17728338, rs999556, and rs3762999 appears to be more extensive in the Chinese than in the European population.32
Figure 3. TNIP1 Polymorphisms Associated with Disease.
In this schematic of the TNIP1 locus, exons are shown in white, grey and red. White and grey exons represent exons that are untranslated and translated, respectively. The red exon represents the ABIN1 AHD4 coding exon. Single nucleotide polymorphisms (variants) found in the TNIP1 locus and associated to autoimmune and inflammatory diseases are indicated by their reference (rs) number and positioned on the TNIP1 locus (References). The majority of these variants are located up or downstream of the TNIP1 coding regions or in intronic regions.
TNIP1 polymorphisms have been implicated in autoimmune disorders, SLE and SSc. Gateva et al. identified the variant, rs7708392, to be associated with SLE patients of European ancestry from the United States and Sweden.19 In replication studies, rs7708392 was also associated with SLE in patients of Japanese and Chinese Han descent.22,25 Han et al. reported association of another TNIP1 variant, rs10036748, with SLE in the Chinese Han population.20 In replication studies, rs10036748 was verified as a risk variant for SLE in the Chinese Han population.25,26 These reports provided evidence for association between TNIP1 polymorphisms and SLE. Yet, these studies were limited in ethnic diversity. Adrianto et al. performed a genetic fine-mapping study in 5 ethnically diverse SLE case-control populations: European, African American, Hispanic, East Asian, and African American Gullah.24 Multiple TNIP1 variants were associated with SLE in persons of European, African American, and Hispanic descent. The presence of these variants with variable pairwise LD suggested the presence of multiple independent SLE-associated haplotypes. Two independent risk haplotypes, H1 and H2, were associated with SLE in patients of European decent that were also present in African American, and Hispanic populations. Zhang et al. identified two risk haplotypes (ATTGCGC, block 1 and GTCCTAT, block 2) to be associated with SLE in the Chinese Han population.25 Of the seven variants in block 2, six were also found in H2. The variants in block 1, however, are independent of those in H1 and H2; demonstrating an independent association between block 1 and SLE in the Chinese Han population. Allanore et al. identified 3 variants (rs3792783, rs2233287, and rs4958881) that confer risk to SSc in persons of European descent.27 After these 3 variants were located in the same LD-block, haplotype block analysis revealed the association of two haplotypes with SSc.27,28 These 3 variants are all found in the H1 haplotype previously associated to SLE, demonstrating similarities in the genetic variation associated to SLE and SSc.24,27
In addition to association with SLE, TNIP1 polymorphisms have been associated with SLE subphenotypes and characteristics. In the Chinese-Han population, a genotype-phenotype analysis of rs10036748 demonstrated association with photosensitivity (P =.01, OR=0.87 [95% CI=0.78–0.97]) and vasculitis (P =.04, OR=1.18 [95% CI=1.01–1.39]).21 In cases of Japanese and Southwestern Chinese ancestry, rs7708392 was significantly associated with renal disorder, antinuclear antibodies, malar rash, and immunologic disorder.22,23 Further support of renal association with TNIP1 polymorphisms was demonstrated in our recent publication. By comparing cases of SLE with and without nephritis (case/case control analysis), strong associations with lupus nephritis were identified at rs7708392 in European Americans (P=3.66×10−4, OR=1.22 [95% CI=1.10–1.37]) and rs4958881 in African Americans (P=8.47×10−3, OR=1.22 [95% CI=1.05–1.42]).9 Comparing SLE cases with nephritis versus healthy controls confirmed the validity of this case/case analysis; revealing a similar association for rs4958881 in African Americans (P=4.43×10−3, OR=1.20) and an even stronger association at rs7708392 in European Americans (P=1.82×10−11, OR=1.44).9 These results indicate that polymorphisms in TNIP1 are associated with nephritis in SLE cases of European and African American decent. It can be concluded that TNIP1 polymorphisms may not only contribute to the development of SLE, but also to its complex subphenotypes and characteristics.
The majority of TNIP1 variants associated with autoimmunity are located up or downstream of the TNIP1 coding regions or in intronic regions; suggesting that they interfere with regulatory elements affecting TNIP1 transcription. Indeed in SLE cases, TNIP1/ABIN-1 mRNA and protein expression is lower in patients harboring TNIP1 risk polymorphisms and haplotypes than controls.24,25 Zhang et al. reported that peripheral blood lymphocytes derived from patients with inactive, mild, moderate, or severe SLE displayed significantly lower levels of TNIP/ABIN1 mRNA and protein compared to healthy controls.25 Adrianto et al. showed that EBV-transformed B cells derived from SLE patients homozygous for either H1 or H2 risk haplotypes displayed reduced expression of TNIP1/ABIN1 mRNA and protein compared to non-risk haplotypes.24 Similar expression level findings were, also, demonstrated in Systemic Sclerosis (SSc) cases. Markedly reduced TNIP1/ABIN1 mRNA and protein expression was observed in both SSc lesional skin tissue and cultured dermal fibroblasts.27 These results link low levels of TNIP1/ABIN1 expression with inflammatory diseases, SLE and SSc. This is in concordance with evidence that elevated levels of NF-κB activity are associated with inflammatory diseases. This TNIP1/ABIN1 expression pattern was not observed in patients suffering from psoriasis and rheumatoid arthritis.29,31 In psoriasis, TNIP1 expression was significantly higher in involved, psoriatic skin biopsies as opposed to uninvolved skin biopsies.29 In rheumatoid arthritis, relatively higher expression levels of TNIP1 mRNA were observed in patients suffering from inflammatory arthritis compared to those with non-inflammatory arthritis.31 Thus, contrary to SLE and SSc reports, higher levels of TNIP1 expression are associated with inflammatory diseases, psoriasis and RA. Since TNIP1 is elevated in these disease states, it seems contradictory to the consensus that enhanced NF-κB activity is associated with inflammatory disease. However, it has been suggested that NF-κB regulates TNIP1 expression at the transcriptional level. Indeed, an NF-κB response element was discovered in the promoter region of TNIP1, and the NF-κB subunit, p65, was shown to bind to this response element.33 Accordingly, overexpression of NF-κB subunits and cytokine-induced stimulation of NF-κB results in the upregulation of TNIP1 mRNA expression.31,33–36 Therefore, the elevated TNIP1 levels found in psoriasis and RA patients may be a direct result of constitutive NF-κB activation, which is present in many autoimmune and inflammatory diseases. Ultimately this suggests a role for ABIN1 in the negative feedback regulation of NF-κB activity.
In conclusion, the genetic association of TNIP1 variants with psoriasis, psoriatic arthritis, SSc, and SLE, together with subphenotype and expression level findings, suggests that TNIP1 plays a critical role in regulating autoimmune disease pathogenesis. Given this association, gene targeting mouse studies are important to characterize the function of ABIN1 in vivo, as well as defining the physiological impact of ABIN1 dysregulation on disease pathogenesis.
TNIP1 Targeting Studies in Mice
Mice harboring TNIP1 deletions and knock-in mutations develop autoimmune phenotypes. Zhou et al. reported that ABIN1 knock-out mice develop a progressive, lupus-like inflammatory disease.37 These mice were generated from ES cells carrying a gene trap cassette in the first intron of TNIP1, preceding the translation-initiating ATG located in the second exon. The ratio of TNIP1−/− pups born alive from heterozygous crossings was lower than Mendelian ratios (4.3% observed versus 25% expected). However, embryonic lethality was influenced by genetic background, as backcrossing from 129S2 ES-cell to C57BL/6 mice resulted in increased TNIP1−/− pups born alive at the F5 generation (10.3% observed versus 25% expected). Initially, live-born TNIP1−/− displayed no macroscopic differences from WT littermates. Overtime, however, they developed a cachectic disease that closely resembled phenotypes demonstrated in patients suffering from SLE. TNIP1−/− mice demonstrated myeloid cell expansion, leukocyte infiltrations in various parenchymatous organs, activated T and B lymphocytes, enlarged spleen and lymph nodes, elevated serum Ig levels, and circulating autoreactive antibodies. Moreover, kidneys developed glomerulonephritis and proteinuria, reflecting renal dysfunction and supporting previous reports of TNIP1 polymorphism association with renal injury.9,22,23 TNIP1−/− mice died prematurely within 4 months, which is reminiscent of premature death associated with SLE patients. Nanda et al. showed that ABIN1 knock-in mice harboring a mutation within the UBAN domain [D485N homologous to human D472N] were incapable of binding polyubiquitin and developed a lupus-like inflammatory disorder.5 Similar to the TNIP1 knockout mouse, [D485N] knock-in mice possessed elevated B cell, granulocyte, and monocyte count, activated T and B lymphocytes, enlarged spleen and lymph nodes, elevated serum Ig levels, circulating autoreactive antibodies, and increased germinal center formation.5,37 Initial analysis of [D485N] kidney sections revealed the presence of immune complex deposits, which initiated the activation of the complement pathway and development of renal disease.5 We further characterized renal abnormalities in these mice and found that [D485N] knock-in mice developed proteinuria and proliferative, immune complex-mediated glomerulonephritis with histologic features of proliferative (class III/IV) human lupus nephritis.9
While ABIN1 knock-out and [D485N] knock-in mice develop similar lupus-like phenotypes, the proposed ABIN1 mechanism of action differs between studies.5,37 Consistent with studies demonstrating the polyubiquitin binding role of ABIN1 in NF-κB inhibition, Nanda et al. showed that in immune cells derived from [D485N] mice, ABIN1 could not bind polyubiquitin and displayed enhanced NF-κB activity via TLR-MyD88 signaling.5 In response to TLR ligands, mutant B lymphocytes also proliferated more rapidly and produced more IL-6 and IL-12 cytokines; hallmarks of NF-κB activation. In addition to NF-κB over-activation, B lymphocytes derived from [D485N] mice showed enhanced activation of mitogen-activated protein kinases (MAPKs; JNK, ERK, and p38α) in response to TLR ligands. Similar signaling results were observed in bone marrow-derived dendritic cells (DCs). Autoimmunity and glomerulonephritis were suppressed in [D485N] mice by crossing to MyD88 deficient mice. This suggests that TLR-MyD88 signaling pathways are needed for the disease phenotype to develop. Therefore, Nanda et al. concluded that the interaction of ABIN1 with polyubiquitin is required to limit the activation of TLR-MyD88 pathways and prevent autoimmunity. Conversely, Zhou et al. proposed an alternative mechanism of ABIN1 in the ABIN1 knockout mouse. In response to TLR stimuli, ABIN1-knock out macrophages exhibit normal regulation of major proinflammatory signaling pathways including NF-κB, JNK, and p38α MAPK pathways.37 However, under the same conditions, selective deregulation of the transcription factor CCAAT/enhancer binding protein β (C/EBPβ) and its target genes colony-stimulating factor 3 (Csf3), S100 calcium-binding protein A8 (S100a8), and nitric oxide synthase, inducible (Nos2) was observed. The gene products of Csf3, S100a8 and Nos2 participate in innate immune cell expansion, host factor-derived inflammation, and cytotoxicity, respectively.38–40 Zhou et al. concluded that ABIN1 loss of function results in a lupus-like inflammatory disease which is attributed, at least in part, to the dysregulation of C/EBPβ activation.
Conditional ABIN1 knock-out findings, further, demonstrate the correlation between ABIN1 dysregulation and autoimmune susceptibility. Callahan et al. generated mice selectively lacking ABIN1 in dendritic cells (DCs), ABIN1fl CD11c-Cre mice.41 Given the importance of DCs in skin immune homeostasis and association between ABIN1 polymorphisms and psoriasis, these authors investigated whether ABIN1 expression in DCs regulates psoriasis susceptibility.29,32,42 Histologic examination of mutant skin sections confirmed epidermal hyperplasia, hypogranulosis, hyperkeratosis, and parakeratosis with neutrophils; stereotypical histologic findings of human psoriasis. Further, treatment of ABIN1fl CD11c-Cre mice with topical TLR ligand markedly increased several hallmark phenotypes of human psoriasis, including prosaic lesions and stimulated IL-23 secretion, Th17 cell differentiation, and neutrophilic inflammation. In concordance with mechanistic findings from Nanda et al., ABIN1−/− bone marrow-derived DCs stimulated with TLR agonists induced exaggerated activation of NF-κB, JNK, and p38.5,41 Moreover, as demonstrated by Nanda et al., TLR-MyD88 signaling is necessary for the exacerbation of disease phenotypes. For, disease phenotypes were suppressed by the DC-specific deletion of MyD88. Overall, this conditional knock-out model shows that ABIN1 expression in DCs is required to limit the activation of TLR-MyD88 pathways and prevent the development of psoriasis.
In addition to its role in autoimmune development, ABIN1 has been implicated in restricting cell death and sustaining embryonic development.13 Oshima et al. generated and characterized the first TNIP1−/− knock-out mouse. These mice were generated by deletion of exons 12–15, abrogating ABIN1 protein expression. They displayed normal Mendelian ratios up to embryonic day 18.5, but then died during late embryogenesis (2.4% born versus 25% expected) from fetal liver apoptosis, anemia, and hypoplasia. TNIP1−/− embryonic fibroblasts were hypersensitive to TNF-induced programmed cell death and, consistent with Zhou et al.’s findings37, lethality was rescued by crossing with mice that did not express the TNFR1 receptor. Thus, ABIN1 sustains embryogenesis by regulating TNF signals and TNF-induced programmed cell death. Mechanistically, ABIN1 was shown to inhibit the TNF-induced interaction between death inducing signaling complex (DISC) proteins: caspase-8 and FADD; preventing caspase-8 cleavage and programmed cell death. It was demonstrated that ABIN1 requires its polyubiquitin binding function to prevent FADD-caspase 8 binding. Therefore, Oshima et al. concluded that ABIN1 requires its ubiquitin binding function to inhibit FADD-caspase 8 association, protect cells against TNF-induced apoptosis, and sustain embryonic development. TNIP1−/− mice generated by Zhou et al., as well as Oshima et al., displayed similar embryonic characteristics; raising the question as to why different findings are reported. While Oshima et al. generated insights on ABIN1’s role in embryonic development it should be noted that the TNIP1−/− live born pups were not further characterized for disease development. Oshima et al. primarily analyzed TNIP1−/− mouse embryonic fibroblasts. Meanwhile, Zhou et al. primarily analyzed immune cells derived from live born TNIP1−/− mice. Therefore, discrepancies observed between these two TNIP1−/− knockout studies may arise from the analysis of different cell types that would reflect the regulation of embryonic development or autoimmune development.
In contrast to ABIN1 observations, ABIN2 and ABIN3-null mice displayed no gross anatomical effects.43,44 ABIN2-null mice were born at the expected Mendelian ratios, were of normal weight, and displayed no signs of inflammation.43 No developmental abnormalities of T or B cell lineages were observed from the thymi, lymph nodes, or spleen of ABIN2-null mice. Further, numbers of macrophages and DC’s from the spleen of ABIN2-null mouse were similar to wild-type. Mechanistically, murine ABIN2 does not regulate NF-κB activity; which may explain the lack of phenotypic observations. 43 ABIN3-null mice were born in normal Mendelian ratios and demonstrated no phenotypic abnormalities.44 This is not surprising, since murine ABIN3 lacks the UBAN domain and, consequentially, is incapable of inhibiting NF-κB activation by proinflammatory stimuli. In opposition to its isoforms, ABIN2 and ABIN3, ABIN1 plays a unique role in the regulation of inflammatory, autoimmune, and embryonic development. The lack of disease phenotype in ABIN2 and ABIN3-deficient mice may be attributed to normal NF-κB regulation.
Concluding Remarks
There is growing evidence indicating that ABIN1 plays an important role in normal tissue homeostasis to prevent autoimmunity and inflammation. Mechanistically, ABIN1 functions as physiological inhibitor of NF-κB in response to TLR, IL-1, CD40, and TNF-mediated signaling. ABIN1 inhibition of other signaling pathways and transcription factors has been reported, but is not well defined and should be explored further. These include inhibition of C/EBPβ, JNK, ERK, and p38 MAPK activity, as well as DISC complex formation. Cell type dependent regulation by ABIN1 is indicated by differential signaling and phenotypic consequences observed between TNIP1−/− derived mouse embryonic fibroblasts and immune cells. Moreover, in a targeted knockout study, mice specifically lacking ABIN1 in dendritic cells develop psoriasis. Additional TNIP1−/− targeted knockout studies have yet to be performed but could further describe ABIN1s cell specific regulatory role. Targeted knockout in B and T lymphocytes would particularly be of interest considering their role in the pathogenesis of autoimmune disorders and the high expression level of ABIN1 in immune cells. At the molecular level, ABIN1 interacts with A20 and polyubiquitinated NF-κB proteins NEMO, IRAK1, and RIP1. ABIN1 mediates A20s interaction with polyubiquitinated NEMO; promoting A20 hydrolase activity, deubiquitination, and NF-κB inhibition.45 Whether ABIN1 mediates A20 function of other polyubiquitinated NF-κB mediator proteins remains unknown. Alternatively, a less defined, A20-independent mechanism is indicated by NF-κB inhibition in the absence of ABIN1’s AHD1 domain. Since familial protein, ABIN2, competes with TRAF6 for ubiquitin chains it is possible that ABIN1 may function independently of A20 by competing with NF-κB mediators for polyubiquitin. Ultimately, to clarify the role of A20 in ABIN1-medated NF-κB inhibition the generation and analysis of double deficient ABIN1/A20 knockout mice may be beneficial. ABIN1 binds to K63-linked and linear polyubiquitin chains through the UBAN domain. ABIN1-UBAN mutants are incapable of restricting TNF and TLR signaling and are associated with murine embryonic lethality, glomerulonephritis, and lupus-like inflammatory disease development. In humans, GWAS-identified ABIN1 polymorphisms are associated with psoriasis, psoriatic arthritis, SLE, and SSc. However, the impact of these polymorphisms on ABIN1 function and polyubiquitin binding is unknown. Therefore, experimentation of patient cells and homologous mutant mice may shed light on how these variants alter the function of ABIN1 and contribute to the pathogenesis of autoimmunity.
Table 2.
ABIN1 Summary.
| Function | Mechanism | Phenotypic Associations | References |
|---|---|---|---|
| Inhibition of NF-κB | Inhibits by an A20 dependent and/or A20 independent mechanism * (See Figure 2) | Lupus-like inflammatory disease Psoriasis Glomerular nephritis |
5, 6, 7, 9, 14, 41 |
| Inhibition of MAPKs (JNK, ERK, and p38α) | Inhibits TLR dependent MAPK activity* Suppresses ERK2 nuclear translocation |
Lupus-like inflammatory disease Psoriasis |
5, 41, 47 |
| Inhibition of C/EBPβ | Inhibits TLR dependent C/EBPβ activity | Lupus-like inflammatory disease | 37 |
| Prevention of apoptosis | Inhibits caspase 8/FADD interaction upon TNFα stimulation* | Embryonic development | 13 |
Requires polyubiquitin binding function
Acknowledgments
Financial Support: This work was supported by funding from NIH/NIAMS R01 AR063124 and NIH/NIAID R21 AI103980.
Footnotes
Conflict of Interest: None
References
- 1.Oeckinghaus A, Ghosh S. The NF-κB Family of Transcription Factors and Its Regulation. Cold Spring Harb Perspect Biol. 2009;1 doi: 10.1101/cshperspect.a000034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wullaert A, Bonnet MC, Pasparakis M. NF-κB in the regulation of epithelial homeostasis and inflammation. Cell Res. 2011;21:146–158. doi: 10.1038/cr.2010.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sun SC, Chang JH, Jin J. Regulation of nuclear factor-κB in autoimmunity. Trends Immunol. 2013;34:282–289. doi: 10.1016/j.it.2013.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pai S, Thomas R. Immune deficiency or hyperactivity-Nf-κb illuminates autoimmunity. J Autoimmun. 2008;31:245–251. doi: 10.1016/j.jaut.2008.04.012. [DOI] [PubMed] [Google Scholar]
- 5.Nanda SK, et al. Polyubiquitin binding to ABIN1 is required to prevent autoimmunity. J Exp Med. 2011;208:1215–1228. doi: 10.1084/jem.20102177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Heyninck K, et al. The zinc finger protein A20 inhibits TNF-induced NF-kappaB-dependent gene expression by interfering with an RIP- or TRAF2-mediated transactivation signal and directly binds to a novel NF-kappaB-inhibiting protein ABIN. J Cell Biol. 1999;145:1471–1482. doi: 10.1083/jcb.145.7.1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heyninck K, Kreike MM, Beyaert R. Structure-function analysis of the A20-binding inhibitor of NF-kappa B activation, ABIN-1. FEBS Lett. 2003;536:135–140. doi: 10.1016/s0014-5793(03)00041-3. [DOI] [PubMed] [Google Scholar]
- 8.Wagner S, et al. Ubiquitin binding mediates the NF-kappaB inhibitory potential of ABIN proteins. Oncogene. 2008;27:3739–3745. doi: 10.1038/sj.onc.1211042. [DOI] [PubMed] [Google Scholar]
- 9.Caster DJ, et al. ABIN1 dysfunction as a genetic basis for lupus nephritis. J Am Soc Nephrol JASN. 2013;24:1743–1754. doi: 10.1681/ASN.2013020148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fukushi M, et al. Identification and cloning of a novel cellular protein Naf1, Nef-associated factor 1, that increases cell surface CD4 expression. FEBS Lett. 1999;442:83–88. doi: 10.1016/s0014-5793(98)01631-7. [DOI] [PubMed] [Google Scholar]
- 11.Favre M, Butticaz C, Stevenson B, Jongeneel CV, Telenti A. High frequency of alternative splicing of human genes participating in the HIV-1 life cycle: a model using TSG101, betaTrCP, PPIA, INI1, NAF1, and PML. J Acquir Immune Defic Syndr 1999. 2003;34:127–133. doi: 10.1097/00126334-200310010-00002. [DOI] [PubMed] [Google Scholar]
- 12.Shiote Y, et al. Multiple splicing variants of Naf1/ABIN-1 transcripts and their alterations in hematopoietic tumors. Int J Mol Med. 2006;18:917–923. [PubMed] [Google Scholar]
- 13.Oshima S, et al. ABIN-1 is a ubiquitin sensor that restricts cell death and sustains embryonic development. Nature. 2009;457:906–909. doi: 10.1038/nature07575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mauro C, et al. ABIN-1 binds to NEMO/IKKgamma and co-operates with A20 in inhibiting NF-kappaB. J Biol Chem. 2006;281:18482–18488. doi: 10.1074/jbc.M601502200. [DOI] [PubMed] [Google Scholar]
- 15.Yuan S, et al. Emergence of the A20/ABIN-mediated inhibition of NF-κB signaling via modifying the ubiquitinated proteins in a basal chordate. Proc Natl Acad Sci U S A. 2014;111:6720–6725. doi: 10.1073/pnas.1321187111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xiong Y, et al. Endotoxin tolerance impairs IL-1 receptor-associated kinase (IRAK) 4 and TGF-beta-activated kinase 1 activation, K63-linked polyubiquitination and assembly of IRAK1, TNF receptor-associated factor 6, and IkappaB kinase gamma and increases A20 expression. J Biol Chem. 2011;286:7905–7916. doi: 10.1074/jbc.M110.182873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jung SM, et al. Smad6 inhibits non-canonical TGF-β1 signalling by recruiting the deubiquitinase A20 to TRAF6. Nat Commun. 2013;4 doi: 10.1038/ncomms3562. [DOI] [PubMed] [Google Scholar]
- 18.Wertz IE, et al. De-ubiquitination and ubiquitin ligase domains of A20 downregulate NF-κB signalling. Nature. 2004;430:694–699. doi: 10.1038/nature02794. [DOI] [PubMed] [Google Scholar]
- 19.Gateva V, et al. A large-scale replication study identifies TNIP1, PRDM1, JAZF1, UHRF1BP1 and IL10 as risk loci for systemic lupus erythematosus. Nat Genet. 2009;41:1228–1233. doi: 10.1038/ng.468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Han JW, et al. Genome-wide association study in a Chinese Han population identifies nine new susceptibility loci for systemic lupus erythematosus. Nat Genet. 2009;41:1234–1237. doi: 10.1038/ng.472. [DOI] [PubMed] [Google Scholar]
- 21.He CF, et al. TNIP1, SLC15A4, ETS1, RasGRP3 and IKZF1 are associated with clinical features of systemic lupus erythematosus in a Chinese Han population. Lupus. 2010;19:1181–1186. doi: 10.1177/0961203310367918. [DOI] [PubMed] [Google Scholar]
- 22.Kawasaki A, et al. Association of TNFAIP3 interacting protein 1, TNIP1 with systemic lupus erythematosus in a Japanese population: a case-control association study. Arthritis Res Ther. 2010;12:R174. doi: 10.1186/ar3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhong H, et al. Replicated associations of TNFAIP3, TNIP1 and ETS1 with systemic lupus erythematosus in a southwestern Chinese population. Arthritis Res Ther. 2011;13:R186. doi: 10.1186/ar3514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Adrianto I, et al. Association of two independent functional risk haplotypes in TNIP1 with systemic lupus erythematosus. Arthritis Rheum. 2012;64:3695–3705. doi: 10.1002/art.34642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang DM, et al. Single-nucleotide polymorphism and haplotypes of TNIP1 associated with systemic lupus erythematosus in a Chinese Han population. J Rheumatol. 2013;40:1535–1544. doi: 10.3899/jrheum.121391. [DOI] [PubMed] [Google Scholar]
- 26.Zuo XB, et al. Variants in TNFSF4, TNFAIP3, TNIP1, BLK, SLC15A4 and UBE2L3 interact to confer risk of systemic lupus erythematosus in Chinese population. Rheumatol Int. 2014;34:459–464. doi: 10.1007/s00296-013-2864-3. [DOI] [PubMed] [Google Scholar]
- 27.Allanore Y, et al. Genome-wide scan identifies TNIP1, PSORS1C1, and RHOB as novel risk loci for systemic sclerosis. PLoS Genet. 2011;7:e1002091. doi: 10.1371/journal.pgen.1002091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bossini-Castillo L, et al. Confirmation of TNIP1 but not RHOB and PSORS1C1 as systemic sclerosis risk factors in a large independent replication study. Ann Rheum Dis. 2013;72:602–607. doi: 10.1136/annrheumdis-2012-201888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nair RP, et al. Genome-wide scan reveals association of psoriasis with IL-23 and NF-kappaB pathways. Nat Genet. 2009;41:199–204. doi: 10.1038/ng.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bowes J, et al. Confirmation of TNIP1 and IL23A as susceptibility loci for psoriatic arthritis. Ann Rheum Dis. 2011;70:1641–1644. doi: 10.1136/ard.2011.150102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gallagher J, et al. Identification of Naf1/ABIN-1 among TNF-α-induced expressed genes in human synoviocytes using oligonucleotide microarrays. FEBS Lett. 2003;551:8–12. doi: 10.1016/s0014-5793(03)00823-8. [DOI] [PubMed] [Google Scholar]
- 32.Sun LD, et al. Association analyses identify six new psoriasis susceptibility loci in the Chinese population. Nat Genet. 2010;42:1005–1009. doi: 10.1038/ng.690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tian B, Nowak DE, Jamaluddin M, Wang S, Brasier AR. Identification of Direct Genomic Targets Downstream of the Nuclear Factor-κB Transcription Factor Mediating Tumor Necrosis Factor Signaling. J Biol Chem. 2005;280:17435–17448. doi: 10.1074/jbc.M500437200. [DOI] [PubMed] [Google Scholar]
- 34.Hinata K, Gervin AM, Jennifer Zhang Y, Khavari PA. Divergent gene regulation and growth effects by NF-κB in epithelial and mesenchymal cells of human skin. Oncogene. 2003;22:1955–1964. doi: 10.1038/sj.onc.1206198. [DOI] [PubMed] [Google Scholar]
- 35.Zhou A, Scoggin S, Gaynor RB, Williams NS. Identification of NF-kappa B-regulated genes induced by TNFalpha utilizing expression profiling and RNA interference. Oncogene. 2003;22:2054–2064. doi: 10.1038/sj.onc.1206262. [DOI] [PubMed] [Google Scholar]
- 36.Németh ZH, et al. cDNA microarray analysis reveals a nuclear factor-kappaB-independent regulation of macrophage function by adenosine. J Pharmacol Exp Ther. 2003;306:1042–1049. doi: 10.1124/jpet.103.052944. [DOI] [PubMed] [Google Scholar]
- 37.Zhou J, et al. A20-binding inhibitor of NF-κB (ABIN1) controls Toll-like receptor-mediated CCAAT/enhancer-binding protein β activation and protects from inflammatory disease. Proc Natl Acad Sci U S A. 2011;108:E998–1006. doi: 10.1073/pnas.1106232108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Metcalf D. Hematopoietic cytokines. Blood. 2008;111:485–491. doi: 10.1182/blood-2007-03-079681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vogl T, et al. Mrp8 and Mrp14 are endogenous activators of Toll-like receptor 4, promoting lethal, endotoxin-induced shock. Nat Med. 2007;13:1042–1049. doi: 10.1038/nm1638. [DOI] [PubMed] [Google Scholar]
- 40.Nagy G, et al. Central role of nitric oxide in the pathogenesis of rheumatoid arthritis and systemic lupus erythematosus. Arthritis Res Ther. 2010;12:210. doi: 10.1186/ar3045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Callahan JA, et al. Cutting Edge: ABIN-1 Protects against Psoriasis by Restricting MyD88 Signals in Dendritic Cells. J Immunol. 2013;191:535–539. doi: 10.4049/jimmunol.1203335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jariwala SP. The role of dendritic cells in the immunopathogenesis of psoriasis. Arch Dermatol Res. 2007;299:359–366. doi: 10.1007/s00403-007-0775-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Papoutsopoulou S, et al. ABIN-2 is required for optimal activation of Erk MAP kinase in innate immune responses. Nat Immunol. 2006;7:606–615. doi: 10.1038/ni1334. [DOI] [PubMed] [Google Scholar]
- 44.Weaver BK, Bohn E, Judd BA, Gil MP, Schreiber RD. ABIN-3: a molecular basis for species divergence in interleukin-10-induced anti-inflammatory actions. Mol Cell Biol. 2007;27:4603–4616. doi: 10.1128/MCB.00223-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Verstrepen L, Carpentier I, Verhelst K, Beyaert R. ABINs: A20 binding inhibitors of NF-kappa B and apoptosis signaling. Biochem Pharmacol. 2009;78:105–114. doi: 10.1016/j.bcp.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 46.Cohen S, Ciechanover A, Kravtsova-Ivantsiv Y, Lapid D, Lahav-Baratz S. ABIN-1 negatively regulates NF-kappaB by inhibiting processing of the p105 precursor. Biochem Biophys Res Commun. 2009;389:205–210. doi: 10.1016/j.bbrc.2009.08.074. [DOI] [PubMed] [Google Scholar]
- 47.Zhang S, et al. A new ERK2 binding protein, Naf1, attenuates the EGF/ERK2 nuclear signaling. Biochem Biophys Res Commun. 2002;297:17–23. doi: 10.1016/s0006-291x(02)02086-7. [DOI] [PubMed] [Google Scholar]