FIG 2.
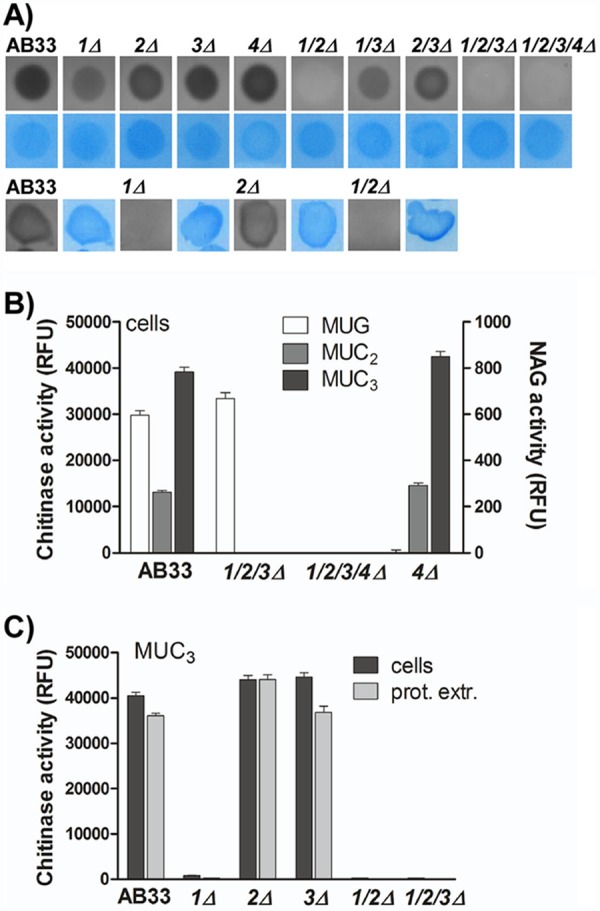
Cts1 and Cts2 contribute to chitin degradation during yeast growth, but only Cts1 is active in filaments. (A) Dot-gel activity assay for chitinase activity with protein extracts of AB33 and chitinase-deficient strains. Chitinase activity is observed as a dark halo. Equal loading was controlled by Coomassie staining (blue). Cts1 and Cts2 contribute to chitin degradation in yeast-like cells (upper), while only Cts1 is active in filaments (lower). (B) Enzymatic activity of yeast cells toward the commercial, fluorogenic substrates for chitinases (MUC2 and MUC3) and N-acetyl-glucosaminidases (NAG) (MUG). Cts4 is the only NAG that does not act on the chitinase substrates, while complete deletions of chitinases do not affect NAG activity. RFU, relative fluorescence units. (C) MUC3 degradation depends on the presence of cts1 but is not affected by deletion of the other chitinases. No difference is detected when intact cells or protein extracts (prot. extr.) are used. Error bars indicate standard deviations from three technical replicates.
