Fig. 2.
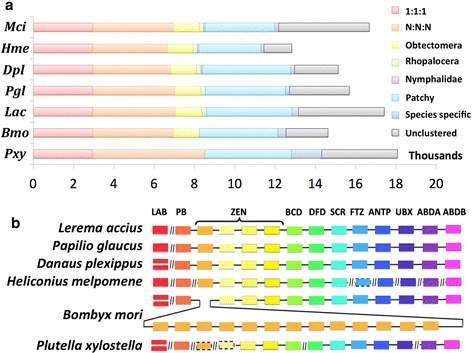
Comparative analysis of Lepidoptera genomes. a Number of different types of orthologs in each Lepidoptera species with published genomes. 1:1:1: single-copy orthologs shared among all species; N:N:N: multiple-copy orthologs shared among all species, i.e. more than one copy in at least one species; Obtectomera: orthologs specific to Obtectomera, i.e. all other six species except Pxy; Rhopalocera: orthologs specific to Rhopalocera, i.e. all other five species except Bmo and Pxy; Nymphalidae: orthologs specific to Nymphalidae, i.e. Dpl, Mci and Hme; Patchy: orthologs that are shared between more than one, but not all species (excluding those belongs to previous categories); Species-specific: specific to only one species and having close homologs within that species; Unclustered: proteins that do not belong to any of the orthologous groups. b Arrangements of Hox genes in Lepidoptera genomes. Orthologs are shown as boxes of the same color; double boxes in the same position indicate gene duplications, dashed-line around a box implies that this gene is missing in the genome assembly but present in the transcriptome; “//” marks the boundaries between different scaffolds
