Fig. 4.
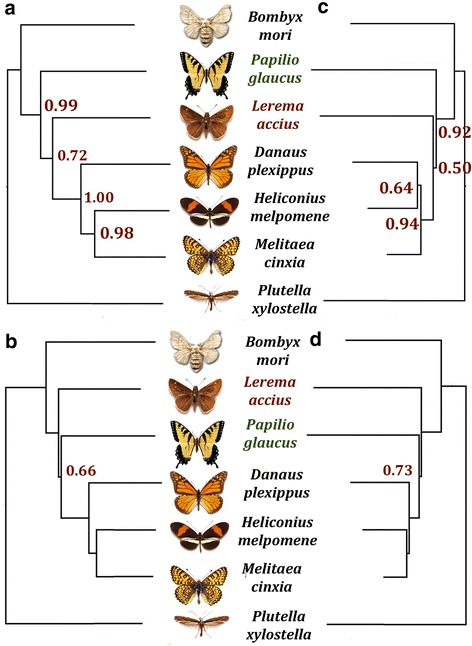
Phylogenetic trees of the butterflies based on whole-genome data. a Majority-rule consensus tree of the maximal likelihood trees constructed by RAxML on 1000 random samples from the concatenated alignment of universal single-copy orthologs. b Neighbor-joining tree based on the frequency of gene rearrangement events between species. c Consensus tree of the better-supported trees inferred from PhyloBayes analyses on 1000 random samples from the concatenated alignment of universal single-copy orthologs. The tree topology was constrained to either of the two reported topologies: (((((Mci, Hme), Dpl), Lac), Pgl), Bmo, Pxy) or (((((Mci, Hme), Dpl), Pgl), Lac), Bmo, Pxy). d Phylogenetic tree using the gene-rearrangement data inferred by PhyloBayes with CAT model
