Figure 8.
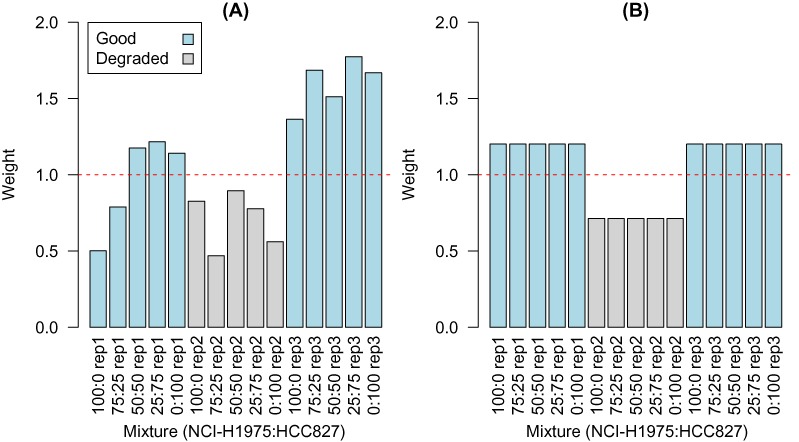
Degraded RNA samples (replicate 2, shaded in grey) from the control experiment are correctly assigned lower weights by the combined ‘voom’ and sample weighting procedure (A), with an average weight of 0.70 across these five samples, compared to an average of 1.28 for the non-degraded samples (replicates 1 and 3, shaded in blue). A similar result is obtained for block weighting (B), with a weight of 0.71 assigned to the five degraded samples versus 1.20 for the remaining samples. When ‘voom’ was combined with sample weighting on the good samples, the weights were equivalent for the replicate 2 samples (1.06) and the remaining samples (1.07, data not shown).
