Figure 1.
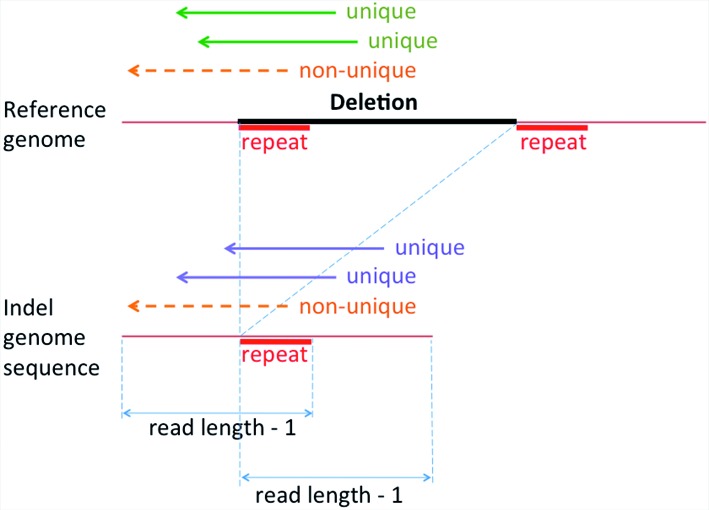
Modified reference genome and calculation of read coverage. Indel genome sequence for the deletion is created by gluing together the two flanking segments of the deletion, after which sequencing reads are aligned simultaneously to both reference and modified sequence. The potential presence of repeat sequences (marked in red) inside/outside the deletion may affect the uniqueness of read alignments: reads that span the entire repeat can be aligned uniquely to either the reference genome (green arrows) or the donor indel sequence (blue arrows), whereas reads not spanning the repeat region (orange dash arrows) will align to both reference and modified sequences equally well. The length of the flanking segments may differ due to read length and repeat length as shown. The coverage of the reference and alternate indel event is calculated as the number of reads that aligned uniquely to each (and not to both).
