Figure 4.
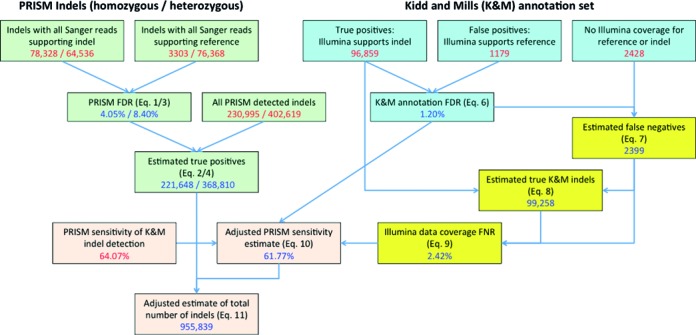
Estimation of the total number of 1–10 bp indels in the Yoruban genome NA18507 using PRISM in combination with BWA read aligner. The workflow involves the estimation of four sets of values: PRISM FDR and the number of true positive indels detected by PRISM (green boxes); the reliability of the reference indel annotation combined from Kidd and Mills sets (via false discovery rate, FDR; blue boxes); the incompleteness of the Illumina read coverage of the reference indels (via false negative rate, FNR; yellow boxes); and the computation of the adjusted sensitivity of indel detection in PRISM, which is used to estimate the overall number of indels in the genome (orange boxes). Each box shows the initial indel counts or pipeline sensitivity (red numbers) as well as the computed estimates (blue numbers) based on the equations indicated in parentheses. The detailed explanation of the equations and the workflow is presented in the Supplementary Materials.
