Figure 6.
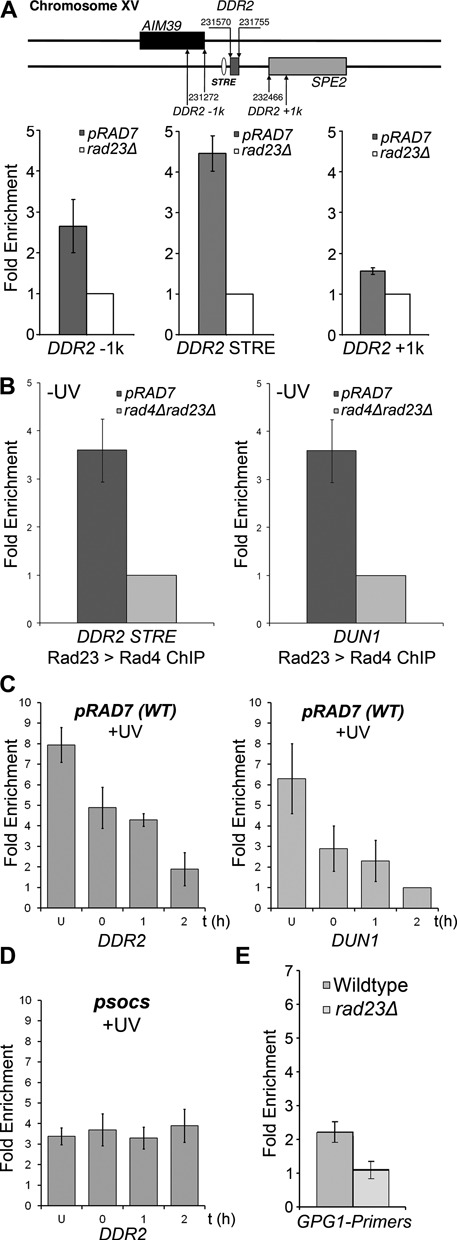
GG-NER E3 ligase dependent Rad4–Rad23 interaction with the promoter regions of the UV induced genes DUN1 and DDR2. (A) ChIP-qPCR data of Rad23 interaction with the DDR2 promoter. Rad23 antibodies were used to immunoprecipitate chromatin bound Rad23. Primer sets covering the STRE and areas 1kb upstream [DDR2–1k] and downstream [DDR2+1k] of the STRE of DDR2 were used to quantify the relative enrichment of Rad23 (top panel). Rad23 protein occupancy at DDR2 in absence of UV irradiation is shown in the lower panel, relative to the ChIP performed on RAD23 deleted cells as a control. (B) Rad23 ChIP was subjected to a second round of IP detecting Rad4 as part of the complex interacting with the DDR2 (right panel) and DUN1 promoters (left panel). (C) Rad4–Rad23 interacts with the DDR2 and DUN1 promoters in response to UV. ChIPs were performed of untreated and UV irradiated chromatin from wildtype cells at different times after UV irradiation. (D) Rad4–Rad23 occupancy at the DDR2 promoter in a GG-NER E3 ligase mutant. As panel (C) but for psocs cells showing no loss of occupancy of the Rad4–Rad23 complex from the DDR2 promoter region. (E) Rad4–Rad23 does not interact with the GPG1 gene promoter. The ChIP-qPCR experiment was performed using Rad23 antibody on wildtype and rad23Δ chromatin. qPCR analysis of the GPG1 gene promoter was performed and shows no enrichment compared to background levels detected in a RAD23 deficient strain. The Rad23 enrichment is relative to the background of the ChIP in RAD23 deletion extracts set to unity. Data shown here are the average of three independent experiments with the standard deviation indicated by the error bars.
