Figure 5.
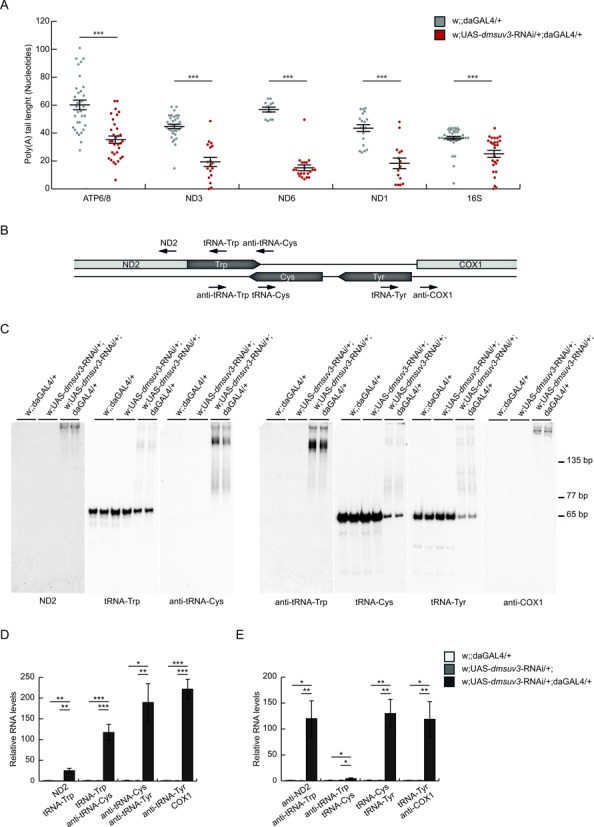
dmsuv3 deficiency leads to a reduction of poly(A) tails and altered processing of mitochondrial tRNAs. (A) Poly(A) tail length in individually sequenced clones after transcript circularization in dmsuv3 KD (w;UAS-dmsuv3-RNAi/+;daGAL4/+) and control (w;;daGAL4/+) larvae at 5 days ael. Mean poly(A) tail length varied between 34 and 60 adenines in control (gray; n ≥ 15) and 11 and 35 adenines in KD (red; n ≥ 24) samples. Data are represented as mean ± SEM (***P < 0.001). (B) Schematic representation of the end-labeled oligonucleotide probes (black arrows) used in northern blot experiments. (C) Northern blot experiments on PAGE separated total RNA using oligonucleotide probes, detailed in (B), against tRNATrp (left panel), tRNACys and tRNATyr (right panel), and their 5′ and 3′ flanking regions in dmsuv3 KD (w;UAS-dmsuv3-RNAi/+;daGAL4/+) and control (w;UAS-dmsuv3-RNAi/+; and w;;daGAL4/+) larvae at 5 days ael. (D) qRT-PCR of the mitochondrial precursor transcripts containing ND2, tRNATrp, anti-tRNACys and COX1 in KD and control larvae at 5 days ael. RP49 transcript was used as endogenous control. (E) qRT-PCR of the mitochondrial precursor transcripts containing tRNACys and tRNATyr in KD and control larvae at 5 days ael. RP49 transcript was used as endogenous control. All data are represented as mean ± SEM (*P < 0.05, **P < 0.01, ***P < 0.001, n = 5).
