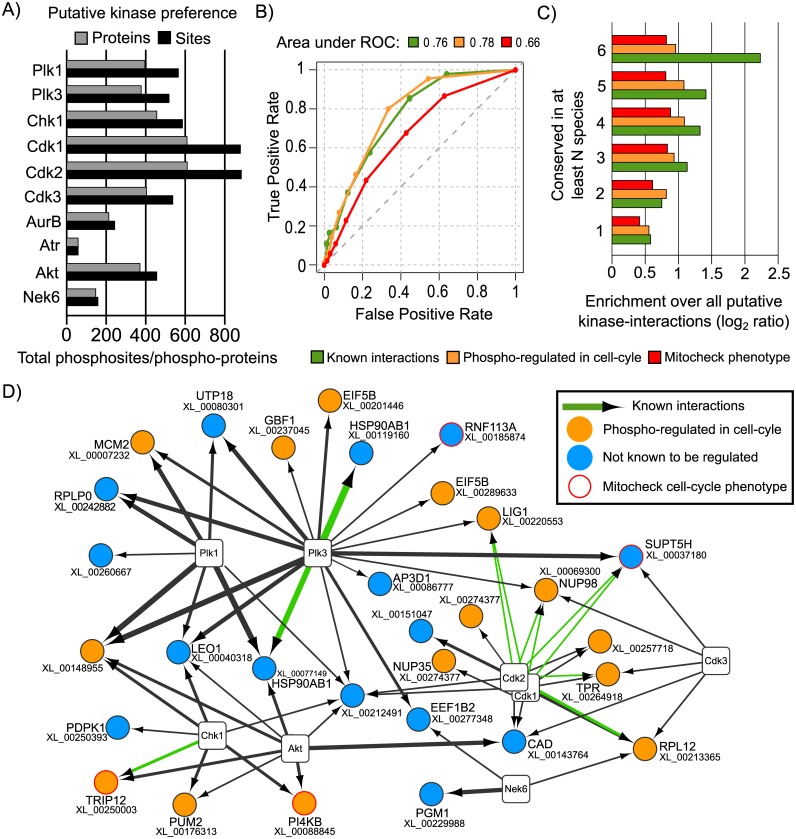
Fig 3. Conservation of putative cell-cycle related kinase interactions is predictive of known and/or cell-cycle related kinase-target interactions.
A) The number of predicted kinase target sites and proteins associated with cell-cycle kinases selected for analysis in X. laevis. We tested if the degree of conservation of kinase-interactions was predictive of known interactions; enriched in proteins that are phospho-regulated in the cell cycle; and genes known to cause cell cycle phenotypes when knocked down. B) ROC curves measuring the accuracy for kinase-interaction predictions. C) Enrichment over random prediction for the 3 tested features. D) Predicted kinase interactions conserved in 7 or more species are shown, highlighting known interactions, proteins phospho-regulated during the cell cycle and genes causing cell cycle phonotypes. The edge thickness is proportional to the degree of conseravtion for the predicted kinase-protein interactions. A list of these interactions is provided in S4 Table.