Abstract
The transcription factor Wilms’ tumor suppressor 1 (WT1) is key to podocyte development and viability; however, WT1 transcriptional networks in podocytes remain elusive. We provide a comprehensive analysis of the genome-wide WT1 transcriptional network in podocytes in vivo using chromatin immunoprecipitation followed by sequencing (ChIPseq) and RNA sequencing techniques. Our data show a specific role for WT1 in regulating the podocyte-specific transcriptome through binding to both promoters and enhancers of target genes. Furthermore, we inferred a podocyte transcription factor network consisting of WT1, LMX1B, TCF21, Fox-class and TEAD family transcription factors, and MAFB that uses tissue-specific enhancers to control podocyte gene expression. In addition to previously described WT1-dependent target genes, ChIPseq identified novel WT1-dependent signaling systems. These targets included components of the Hippo signaling system, underscoring the power of genome-wide transcriptional-network analyses. Together, our data elucidate a comprehensive gene regulatory network in podocytes suggesting that WT1 gene regulatory function and podocyte cell-type specification can best be understood in the context of transcription factor-regulatory element network interplay.
Keywords: podocyte, gene expression, genetics and development, gene transcription, glomerular disease, glomerulus
Renal filtration barrier diseases are a leading cause of end-stage renal failure. Most disorders affect the glomerular podocytes, terminally differentiated and highly polarized cells that require tightly controlled signaling to maintain their integrity, viability, and function.1 Although the most abundant podocyte transcription factor (TF), Wilms’ tumor suppressor 1 (WT1), was identified as a key podocyte regulator,2–4 the WT1-controlled transcriptional podocyte network is poorly understood. How WT1 conveys its cell-type specificity and in which context WT1 acts as a TF in podocytes is unclear. We conducted a genome and transcriptome-wide analysis of the WT1-dependent gene regulatory network in podocytes on the basis of next-generation sequencing of chromatin immunoprecipitation (ChIPseq) and mRNA samples (RNAseq) from mouse glomeruli. We performed ChIPseq for WT1 on kidneys of wild-type mice and analyzed the datasets along ENCODE guidelines.5 The dataset was on the basis of three high quality replicates (Supplemental Table 1) yielding 14,863 reproducible binding sites (peaks) within the podocyte genome (Figure 1A, Supplemental Figure 1, A and B). Several of those peaks were validated by ChIP followed by quantitative PCR (Supplemental Figure 1C). WT1 peaks showed high cross-species conservation scores and harbored a canonical WT1 binding motif at their centers, which was highly similar to WT1 motifs derived from previously published WT1 ChIP-on-chip and ChIPseq experiments in other tissues (Figure 1, B–D).6,7 Basically all WT1 target genes in podocytes known to date were confirmed as bound by WT1.8–11 These analyses indicated that we acquired an extensive and robust representation of genome-wide podocyte WT1 binding sites from living animals.
Figure 1.
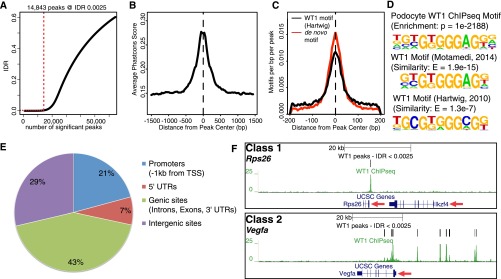
WT1 ChIPseq reproducibly identifies WT1 binding at two different classes of podocyte target genes. (A) Identification of reproducible WT1 binding sites (peaks) by IDR (irreproducible discovery rate) analysis. Red dotted lines indicate the number of peaks at the given IDR threshold. 14,843 peaks are identified at an IDR of 0.25%. (B) Distribution of average cross-species conservation scores (Phastcons) within WT1 peaks indicates increased conservation in peak centers and identification of conserved WT1 binding sites. Dotted line represents the peak center. (C) Localization of a podocyte and previously published WT1-binding motif within WT1 peaks shows enrichment of WT1 motifs in peak centers, confirming the validity of WT1 ChIPseq. (D) Podocyte-derived WT1 binding motif compared with previously published WT1 motifs identified in ChIP-on-chip (ref. 6) and ChIPseq (ref. 7) experiments in embryonic kidneys. The de novo podocyte motif yields highly significant enrichment and similarity scores. (E) Pie chart representing the distribution of WT1 peaks within the annotated genome. One-third of WT1 peaks were located in promoters and 5′-UTRs; most of the sites was found in genic/ intergenic loci. (F) Genome browser plots of WT1 binding sites, gene location, and cross-species conservation showing representative examples for the two different classes of WT1 target genes. Class 1 targets are bound exclusively close to their TSSs as shown for Rps26, whereas class 2 genes have additional binding sites in genic/intergenic regions within the genes’ cis-regulatory domains as shown for Vegfa. Red arrows: direction of transcription.
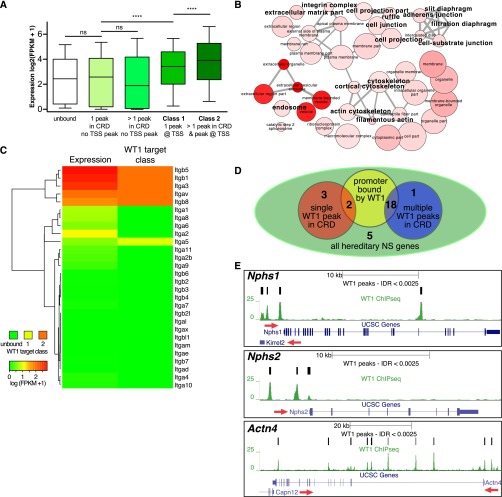
We next analyzed binding patterns of WT1 with respect to the annotated genome. One-third of the WT1 peaks were located in promoters and 5′-UTRs close to transcription start sites (TSSs). However, most peaks were found within genic and intergenic regions (Figure 1E), suggesting a significant relevance of nonpromoter WT1 binding events. Further characterization of WT1 binding patterns revealed two classes of WT1 target genes (Figure 1F): class 1 genes were exclusively bound at their TSSs, whereas class 2 genes were bound at their TSSs and at additional sites within genic or intergenic regions in the genes’ cis-regulatory domains. To elucidate whether these different binding patterns had functional implications, we generated RNAseq data from murine FACS-sorted podocytes (Supplemental Figure 2) and correlated average mRNA expression levels to WT1 binding status (Figure 2A). Interestingly, the average expression level of class 1 genes was higher than the expression level of unbound genes, with average expression of class 2 genes further exceeding that of class 1. Together, these data suggested that WT1 has a predominant transcriptional activating effect in podocytes. The scope of this regulation extends far beyond the short list of previously described target genes and requires concerted TSSs and distal WT1 binding event action.
Figure 2.
WT1 activates a podocyte-specific transcriptome and targets most hereditary podocytopathy genes. (A) Box plots correlating WT1 target gene classes and binding patterns to podocyte gene expression levels by RNAseq. The presence of WT1 binding sites at target gene TSSs (class 1) is associated with higher absolute mRNA expression level average when compared with the average expression level of genes lacking a WT1 binding TSS. The average absolute expression level of genes positive for genic/intergenic WT1 peaks in addition to a WT1-bound TSS (class 2) further exceeds the average level of class 1 gene expression. The average expression level of genes positive for WT1 peaks within their cis-regulatory domains, but without a peak close to their TSSs, did not differ from unbound genes, suggesting that the predominant effect of WT1 in podocytes is transcription activation. ns, not significant. ****P<0.001. (B) GO enrichment network for cellular components derived from class 2 WT1 target genes shows enrichment of major podocyte components. Color intensity increases with significance, node size represents abundance of term in database, and edge strength indicates similarity. A larger font size indicates terms with particular relevance to podocytes. (C) Heat map and clustering analysis correlating expression levels of the integrin gene family to WT1 target classes. A class 2 binding pattern specifically identifies integrins expressed in podocytes. (D) Venn diagram summarizing binding patterns at hereditary podocytopathy genes. Eighteen genes are bound in a class 2 pattern, with a further two genes in a class 1 pattern. (E) Genome browser plots of WT1 binding sites, gene location, and conservation showing class 2 binding patterns at the three key hereditary NS/FSGS genes, Nphs1, Nphs2, and Actn4. Red arrows indicate direction of transcription.
We next analyzed if class 1 and class 2 targets differed functionally using gene ontology (GO) enrichment approaches. General pathways of protein metabolism and transcription enriched among class 1 genes (Supplemental Figure 3, Supplemental Table 2), whereas class 2 genes constituted a toolkit to specifically assemble podocytes. For instance, GO terms for key podocyte components, such as the slit diaphragm, actin cytoskeleton, basement membrane, and focal adhesions, were enriched in a highly specific manner among the class 2 target genes (Figure 2B). This specificity was illustrated by analyzing gene families such as the integrin family. A clustering analysis of all 28 murine integrin-gene mRNA expression levels with their WT1 binding status revealed that exclusively the five class 2 bound integrins were expressed at high levels in podocytes (Figure 2C). Similar results were obtained for the collagen and laminin gene families (Supplemental Figure 4). These findings support the conclusion that a class 2 WT1 binding pattern specifies key elements of the podocyte-specific transcriptome, suggesting that the interplay between podocyte TFs with WT1-bound enhancers may confer cell-type specification.
WT1 mutations in humans either result in developmental defects or in steroid-resistant nephrotic syndrome (NS) with FSGS.12–14 Therefore, we next analyzed if class 2 binding pattern genes could also explain the association of WT1 mutations with hereditary podocytopathies. WT1 bound to 18 of the 31 known podocytopathy genes in a class 2 pattern, including many prevalent genes that cause hereditary FSGS and NS when mutated (Figure 2, D and E, Supplemental Table 3).12 Hence, WT1 mutations with subsequent altered transcriptional regulation of hereditary podocytopathy genes may contribute to the pathogenesis of FSGS and NS. We suggest that the analyses of mRNA expression levels together with WT1 binding status provides a utilitarian transcriptional network identifying mechanisms relevant to podocyte biology.
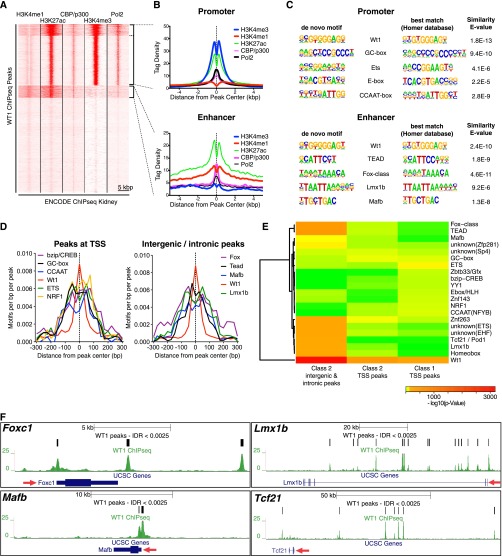
Despite these key WT1 functions, podocytes probably rely on numerous TFs to control gene expression. Indeed, MAFB/Kreisler, TCF21/POD1, LMX1B, and the FoxC-class TFs have already been shown to be of relevance in podocytes.15–19 Frequently, networks of TFs cooperatively control gene expression by binding in TF modules to enhancers. These tissue-specific DNA-regulatory elements are located distally from the TSS.20 We therefore characterized cooperative TF action in podocytes by categorizing WT1 peaks on the basis of chromatin signatures and analyzed DNA-motif enrichment in such peak categories. We performed a clustering analysis of WT1 peaks with ENCODE ChIPseq data obtained from whole mouse kidneys for histone modifications, the enhancer-binding protein p300, and RNA-polymerase 2 (Pol2) (Figure 3A).21 In line with our previous findings, this analysis revealed a promoter cluster comprising one-third of the peaks (Figure 3, A and B). More importantly, we also identified a cluster of approximately 1000 peaks harboring an active enhancer chromatin signature,22 confirming the fact that WT1 binds to enhancers in podocytes (Figure 3, A and B). The small size of this cluster can be explained by the use of whole kidney data for clustering, which no longer provides the cell-type specificity typical of enhancers. When analyzing DNA-motif enrichment independently, the promoter cluster was predominantly enriched in GC boxes, which are used to efficiently assemble the transcription machinery in TATA-less promoters (Figure 3C).23 However, the enhancer cluster revealed binding sites for TF families with established functions in podocytes, such as Fox-class TFs, LMX1B, and MAFB. The motifs for these TFs accumulated in close vicinity of WT1 peak centers (Figure 3D), suggesting that they can be cobound as part of a WT1-TF protein complex. Because clustering on the basis of histone modifications did not differentiate between class 1 and class 2 sites at TSS, we repeated the enrichment analyses with peak sets categorized according to WT1 target gene class, gene expression levels, and location (Figure 3E, Supplemental Figures 5 and 6). This analysis revealed differences in promoter DNA-motif utilization between class 1 and class 2 genes, with predominant enrichment of GC boxes and Znf263 sites in class 2 promoter peaks, whereas class 1 promoter peaks were enriched in Ets-class, CCAAT, and NRF1 motifs (Figure 3E, Supplemental Figure 5). Interestingly, NRF1 activates transcription of genes involved in mitochondrial function, protein catabolism, and cell-cycle control, corroborating the results of our GO enrichment analysis for class 1 peaks.24,25 The analysis of class 2 nonpromoter peaks confirmed a highly significant enrichment of Homeobox, Tcf-family, MAFB, and Fox-class sites, suggesting that these factors may cooperate with WT1 to activate podocyte-specific enhancers and constitute a podocyte-TF network (Figure 3E). Indeed, as frequently found in such networks, WT1 targeted these TF genes to activate their expression in podocytes (Figure 3F). Furthermore, previously described enhancers conferring podocyte-specific gene expression and the TFs bound to these enhancers were recapitulated by our ChIPseq data and motif analysis (Supplemental Figure 7).8,16,19 Reanalysis of previously published WT1 ChIPseq data from embryonic kidneys, where WT1 is expressed in nephron progenitor cells and podocytes, confirmed the tissue specificity of this network (Supplemental Figure 8).7 The fractional overlap of embryonic kidney and podocyte peaks was lowest among distal peaks (Supplemental Figure 8A), with distal peaks specific to embryonic kidneys being exclusively enriched for GO terms in kidney development. In contrast, distal peaks common to podocytes and embryonic kidneys also revealed GO terms with relevance to podocytes (Supplemental Figure 8B). Motif enrichment analysis of the two peak sets showed differences in motif usage and therefore associated TF modules (Supplemental Figure 8C). In summary, these findings suggest a podocyte-specific cooperative network of WT1 with further TFs, integrating WT1 binding with Fox, LMX1B, MAFB, and TCF21 functions (Supplemental Figure 9).
Figure 3.
WT1 enriches a podocyte-specific TF module at its non-TSS binding sites. (A) Clustering analysis of WT1 peaks (rows) with ENCODE kidney ChIPseq tag intensities for indicated histone modifications and TFs (columns) reveals clusters with promoter and enhancer chromatin signatures. The promoter clusters (top) are enriched in H3K4me3, H3K27ac, and Pol2, whereas an active enhancer cluster (middle) is enriched in H3K4me1, p300, and H3K27ac. The bottom cluster shows a histone signature similar to the enhancer cluster, however at much lower density values (profile not shown in B). (B) Histone modification tag density profiles within WT1 peaks for clusters identified in A. Dotted lines indicate peak centers. In the promoter cluster, signal intensities for H3K4me3 and Pol2 are higher than the intensities for H3K4me1 and p300, respectively. The opposite is true for the enhancer cluster. (C) Top five DNA binding motifs identified in separate motif enrichment analyses on WT1 peak clusters identified in A. Only motifs at a false discovery rate (FDR) <0.01 are included. Manually curated best matching motifs from public databases are shown on the right with a similarity E value. Promoters are enriched in motifs generally found close to TSS (Ets-class TF, E box, GC box, CCAAT box), whereas enhancers harbor motifs for TFs with established significance in podocytes (Fox-class TFs, Lmx1b, Mafb). WT1 motifs are highly enriched in promoters and enhancers, confirming that DNA in both types of elements can be directly bound by WT1. (D) Localization analysis of motifs identified in C within WT1 peaks. Dotted line indicates peak centers. Motifs identified in C are enriched close to WT1-bound sites, suggesting a potential for cooperative binding. (E) Heat map showing nonscaled enrichment P values of motifs identified in C and from further analyses (Supplemental Figures 5 and 6). Peak sets corresponding to different target gene classes and different WT1 binding positions (TSS versus genic/intergenic) are analyzed separately in columns. All peak sets are enriched in WT1 motifs. The genic/intergenic class 2 set is enriched in motifs for TFs with known functions in podocytes (Fox-class, Tcf21, Lmx1b, Mafb) and further motifs. The TSS peak sets show different motif usage depending on their WT1 target class. (F) Genome browser plots of WT1-bound TF genes with established relevance and expression in podocytes. Expression, WT1 binding, and motif enrichment of these TFs at WT1 enhancer peaks suggests integration of these TFs in a podocyte TF network (see Supplemental Figure 9 for schematic representation). Red arrows indicate direction of transcription.
Finally, we used our WT1 ChIPseq and RNAseq datasets to derive information on novel pathways with relevance to podocytes. We were intrigued to find enrichment of motifs for TEAD TFs, downstream effectors of hippo signaling, in the vicinity of WT1 ChIPseq peaks (Figure 3, C–E). These TEAD motifs were not enriched in WT1 peaks exclusive to embryonic kidneys and therefore appear to be podocyte specific (Supplemental Figure 8C). The hippo pathway plays significant roles in the control of organ size, apoptosis, and the regulation of cell-matrix adhesions in various organ systems, including the developing kidney.26–28 Although an antiapoptotic function of the hippo downstream effector, Yes-associated protein, in immortalized murine podocyte cell lines has been suggested,29 the in vivo relevance of the hippo pathway in podocytes has not been shown. Interestingly, the hippo signaling cascade was among the top 40 enriched GO terms for biologic processes (Figure 4A). We therefore analyzed expression and WT1 binding status of core components of the hippo-signaling cascade. Strikingly, we found not only the transcriptionally active nuclear components, but also gene coding for almost the entire pathway to be expressed in podocytes and bound by WT1 (Figure 4, B and C). Furthermore, when performing GO enrichment analysis on TEAD-motif positive WT1 peaks against a background of all WT1 peaks, a significant enrichment of terms relating to cell-matrix adhesions and the closely connected actin cytoskeleton organization were identified (Figure 4D). These findings indicate an in vivo role for hippo signaling in podocytes in cooperation with WT1 and open entirely new alleys for investigation in podocyte biology.
Figure 4.
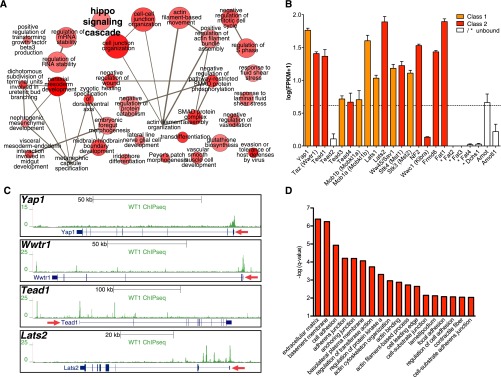
WT1 ChIPseq suggests the hippo pathway to be relevant in podocytes in vivo. (A) GO enrichment network for biologic processes from the top 40 enriched GO terms among WT1 binding sites as determined by the GREAT algorithm. The hippo signaling cascade is identified as enriched (font size arbitrarily increased). (B) Expression levels of core hippo signaling pathway genes in podocytes in vivo from RNAseq experiments (n=3, mean±SEM). WT1 target class is color coded and indicated by asterisks for nonexpressed genes. Dashed line indicates the threshold to call genes expressed versus not expressed as derived from Supplemental Figure 2B. All core constituents of the hippo pathway are expressed in podocytes. (C) Genome browser plots of peaks at WT1-bound hippo pathway genes. Red arrow indicates direction of transcription. (D) GO enrichment analysis (GREAT) of terms enriched at least 1.5-fold among TEAD-binding motif containing WT1 peaks against a background of all WT1 peaks. Redundant terms were manually removed. Terms related to hippo signaling functions, such as cell-matrix interactions and actin cytoskeleton regulation, are overrepresented.
In conclusion, we provide a comprehensive dataset based on an extensive analysis of WT1-directed transcriptional networks in podocytes in vivo. This new database may serve as the basis for further analyses of transcriptional regulation in healthy and diseased podocytes, thereby providing perspectives for additional translational research.
Concise Methods
ChIP was carried out on pooled samples of six CD-1 wild-type mice aged 3 weeks (n=3) using an anti-WT1 antibody (C19; Santa Cruz Biotechnology). Enrichment of WT1-bound DNA was confirmed by ChIP-quantitative PCR on a Cepheid SmartCycler using the promoter of Jmjd1a/Kdm3b as a positive control and an intron of Gapdhs as a negative control. Sequencing libraries were constructed using the SPRYworks system. For RNAseq analysis, GFP was conditionally expressed in podocytes by crossing Nphs2-Cre mice with R26-mTmG reporter mice, and podocytes were isolated by FACS as described.30 Total RNA was extracted from GFP-positive cells using a miRNeasy RNA extraction kit (Qiagen), RNA quality was assayed on a TapeStation RNA Analyzer (Agilent), and sequencing libraries were constructed. All sequencing was carried out on an Illumina HiSeq system.
For bioinformatic analyses, various software was used, including Eland and Bowtie mapping software; SPP for peak calling; IDR for reproducibility analysis; HOMER for motif analysis; GREAT, GOrilla, DAVID, REViGO, and Cytoscape for GO enrichment analyses; and seqMINER for clustering. Raw and processed sequencing data have been deposited in GEO (GSE64063) (accessible at http://www.ncbi.nlm.gov/geo/query/acc.cgi?acc=GSE64063). For details refer to the Detailed Methods section of the Supplemental Material.
All mouse studies were carried out in accordance with the policies of the Institutional Animal Care and Use Committee at Boston Children’s Hospital.
Disclosures
None.
Acknowledgments
We would like to thank Argyris Papantonis and Friedrich C. Luft for critical reading of the manuscript.
M.K. was a recipient of scholarships by the German Research Foundation (KA-3217/2-1), the German Hypertension Society, and CECAD Cologne. J.A.K. received grant support from R01-DK087794-A1. T.B. received grant support by the German Research Foundation (BE2212 and SFB635). M.O.L. was supported by a KoelnFortune scholarship.
Parts of this work were presented in abstract format at the 10th International Podocyte Conference 2014, the Annual Meeting of the German Society of Nephrology 2014, and the 2014 Kidney Week of the American Society of Nephrology.
Footnotes
Published online ahead of print. Publication date available at www.jasn.org.
This article contains supplemental material online at http://jasn.asnjournals.org/lookup/suppl/doi:10.1681/ASN.2014090940/-/DCSupplemental.
References
- 1.Brinkkoetter PT, Ising C, Benzing T: The role of the podocyte in albumin filtration. Nat Rev Nephrol 9: 328–336, 2013 [DOI] [PubMed] [Google Scholar]
- 2.Chau YY, Brownstein D, Mjoseng H, Lee WC, Buza-Vidas N, Nerlov C, Jacobsen SE, Perry P, Berry R, Thornburn A, Sexton D, Morton N, Hohenstein P, Freyer E, Samuel K, van’t Hof R, Hastie N: Acute multiple organ failure in adult mice deleted for the developmental regulator Wt1. PLoS Genet 7: e1002404, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhou L, Li Y, He W, Zhou D, Tan RJ, Nie J, Hou FF, Liu Y: Mutual antagonism of Wilms’ tumor 1 and β-catenin dictates podocyte health and disease. J Am Soc Nephrol 2014, in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Essafi A, Webb A, Berry RL, Slight J, Burn SF, Spraggon L, Velecela V, Martinez-Estrada OM, Wiltshire JH, Roberts SG, Brownstein D, Davies JA, Hastie ND, Hohenstein P: A wt1-controlled chromatin switching mechanism underpins tissue-specific wnt4 activation and repression. Dev Cell 21: 559–574, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Landt SG, Marinov GK, Kundaje A, Kheradpour P, Pauli F, Batzoglou S, Bernstein BE, Bickel P, Brown JB, Cayting P, Chen Y, DeSalvo G, Epstein C, Fisher-Aylor KI, Euskirchen G, Gerstein M, Gertz J, Hartemink AJ, Hoffman MM, Iyer VR, Jung YL, Karmakar S, Kellis M, Kharchenko PV, Li Q, Liu T, Liu XS, Ma L, Milosavljevic A, Myers RM, Park PJ, Pazin MJ, Perry MD, Raha D, Reddy TE, Rozowsky J, Shoresh N, Sidow A, Slattery M, Stamatoyannopoulos JA, Tolstorukov MY, White KP, Xi S, Farnham PJ, Lieb JD, Wold BJ, Snyder M: ChIP-seq guidelines and practices of the ENCODE and modENCODE consortia. Genome Res 22: 1813–1831, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hartwig S, Ho J, Pandey P, Macisaac K, Taglienti M, Xiang M, Alterovitz G, Ramoni M, Fraenkel E, Kreidberg JA: Genomic characterization of Wilms’ tumor suppressor 1 targets in nephron progenitor cells during kidney development. Development 137: 1189–1203, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Motamedi FJ, Badro DA, Clarkson M, Lecca MR, Bradford ST, Buske FA, Saar K, Hübner N, Brändli AW, Schedl A: WT1 controls antagonistic FGF and BMP-pSMAD pathways in early renal progenitors. Nat Commun 5: 4444, 2014 [DOI] [PubMed] [Google Scholar]
- 8.Guo G, Morrison DJ, Licht JD, Quaggin SE: WT1 activates a glomerular-specific enhancer identified from the human nephrin gene. J Am Soc Nephrol 15: 2851–2856, 2004 [DOI] [PubMed] [Google Scholar]
- 9.Palmer RE, Kotsianti A, Cadman B, Boyd T, Gerald W, Haber DA: WT1 regulates the expression of the major glomerular podocyte membrane protein Podocalyxin. Curr Biol 11: 1805–1809, 2001 [DOI] [PubMed] [Google Scholar]
- 10.Schumacher VA, Schlötzer-Schrehardt U, Karumanchi SA, Shi X, Zaia J, Jeruschke S, Zhang D, Pavenstädt H, Drenckhan A, Amann K, Ng C, Hartwig S, Ng KH, Ho J, Kreidberg JA, Taglienti M, Royer-Pokora B, Ai X: WT1-dependent sulfatase expression maintains the normal glomerular filtration barrier. J Am Soc Nephrol 22: 1286–1296, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hanson J, Gorman J, Reese J, Fraizer G: Regulation of vascular endothelial growth factor, VEGF, gene promoter by the tumor suppressor, WT1. Front Biosci 12: 2279–2290, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lovric S, Fang H, Vega-Warner V, Sadowski CE, Gee HY, Halbritter J, Ashraf S, Saisawat P, Soliman NA, Kari JA, Otto EA, Hildebrandt F, Nephrotic Syndrome Study Group : Rapid detection of monogenic causes of childhood-onset steroid-resistant nephrotic syndrome. Clin J Am Soc Nephrol 9: 1109–1116, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Iijima K, Someya T, Ito S, Nozu K, Nakanishi K, Matsuoka K, Ohashi H, Nagata M, Kamei K, Sasaki S: Focal segmental glomerulosclerosis in patients with complete deletion of one WT1 allele. Pediatrics 129: e1621–e1625, 2012. [DOI] [PubMed] [Google Scholar]
- 14.Chernin G, Vega-Warner V, Schoeb DS, Heeringa SF, Ovunc B, Saisawat P, Cleper R, Ozaltin F, Hildebrandt F, Members of the GPN Study Group : Genotype/phenotype correlation in nephrotic syndrome caused by WT1 mutations. Clin J Am Soc Nephrol 5: 1655–1662, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Burghardt T, Kastner J, Suleiman H, Rivera-Milla E, Stepanova N, Lottaz C, Kubitza M, Böger CA, Schmidt S, Gorski M, de Vries U, Schmidt H, Hertting I, Kopp J, Rascle A, Moser M, Heid IM, Warth R, Spang R, Wegener J, Mierke CT, Englert C, Witzgall R: LMX1B is essential for the maintenance of differentiated podocytes in adult kidneys. J Am Soc Nephrol 24: 1830–1848, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morito N, Yoh K, Ojima M, Okamura M, Nakamura M, Hamada M, Shimohata H, Moriguchi T, Yamagata K, Takahashi S: Overexpression of Mafb in podocytes protects against diabetic nephropathy. J Am Soc Nephrol 25: 2546–2557, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maezawa Y, Onay T, Scott RP, Keir LS, Dimke H, Li C, Eremina V, Maezawa Y, Jeansson M, Shan J, Binnie M, Lewin M, Ghosh A, Miner JH, Vainio SJ, Quaggin SE: Loss of the podocyte-expressed transcription factor Tcf21/Pod1 results in podocyte differentiation defects and FSGS. J Am Soc Nephrol 25: 2459–2470, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miner JH, Morello R, Andrews KL, Li C, Antignac C, Shaw AS, Lee B: Transcriptional induction of slit diaphragm genes by Lmx1b is required in podocyte differentiation. J Clin Invest 109: 1065–1072, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.He B, Ebarasi L, Zhao Z, Guo J, Ojala JR, Hultenby K, De VS, Betsholtz C, Tryggvason K: Lmx1b and FoxC combinatorially regulate podocin expression in podocytes. J Am Soc Nephrol 25: 2764–2777, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thurman RE, Rynes E, Humbert R, Vierstra J, Maurano MT, Haugen E, Sheffield NC, Stergachis AB, Wang H, Vernot B, Garg K, John S, Sandstrom R, Bates D, Boatman L, Canfield TK, Diegel M, Dunn D, Ebersol AK, Frum T, Giste E, Johnson AK, Johnson EM, Kutyavin T, Lajoie B, Lee BK, Lee K, London D, Lotakis D, Neph S, Neri F, Nguyen ED, Qu H, Reynolds AP, Roach V, Safi A, Sanchez ME, Sanyal A, Shafer A, Simon JM, Song L, Vong S, Weaver M, Yan Y, Zhang Z, Zhang Z, Lenhard B, Tewari M, Dorschner MO, Hansen RS, Navas PA, Stamatoyannopoulos G, Iyer VR, Lieb JD, Sunyaev SR, Akey JM, Sabo PJ, Kaul R, Furey TS, Dekker J, Crawford GE, Stamatoyannopoulos JA: The accessible chromatin landscape of the human genome. Nature 489: 75–82, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shen Y, Yue F, McCleary DF, Ye Z, Edsall L, Kuan S, Wagner U, Dixon J, Lee L, Lobanenkov VV, Ren B: A map of the cis-regulatory sequences in the mouse genome. Nature 488: 116–120, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, Ye Z, Lee LK, Stuart RK, Ching CW, Ching KA, Antosiewicz-Bourget JE, Liu H, Zhang X, Green RD, Lobanenkov VV, Stewart R, Thomson JA, Crawford GE, Kellis M, Ren B: Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature 459: 108–112, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Anish R, Hossain MB, Jacobson RH, Takada S: Characterization of transcription from TATA-less promoters: Identification of a new core promoter element XCPE2 and analysis of factor requirements. PLoS One 4: e5103, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tsuchiya Y, Taniguchi H, Ito Y, Morita T, Karim MR, Ohtake N, Fukagai K, Ito T, Okamuro S, Iemura S, Natsume T, Nishida E, Kobayashi A: The casein kinase 2-nrf1 axis controls the clearance of ubiquitinated proteins by regulating proteasome gene expression. Mol Cell Biol 33: 3461–3472, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Satoh J, Kawana N, Yamamoto Y: Pathway analysis of ChIP-Seq-based NRF1 target genes suggests a logical hypothesis of their involvement in the pathogenesis of neurodegenerative diseases. Gene Regul Syst Bio 7: 139–152, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aragona M, Panciera T, Manfrin A, Giulitti S, Michielin F, Elvassore N, Dupont S, Piccolo S: A mechanical checkpoint controls multicellular growth through YAP/TAZ regulation by actin-processing factors. Cell 154: 1047–1059, 2013 [DOI] [PubMed] [Google Scholar]
- 27.Dupont S, Morsut L, Aragona M, Enzo E, Giulitti S, Cordenonsi M, Zanconato F, Le Digabel J, Forcato M, Bicciato S, Elvassore N, Piccolo S: Role of YAP/TAZ in mechanotransduction. Nature 474: 179–183, 2011 [DOI] [PubMed] [Google Scholar]
- 28.Reginensi A, Scott RP, Gregorieff A, Bagherie-Lachidan M, Chung C, Lim DS, Pawson T, Wrana J, McNeill H: Yap- and Cdc42-dependent nephrogenesis and morphogenesis during mouse kidney development. PLoS Genet 9: e1003380, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Campbell KN, Wong JS, Gupta R, Asanuma K, Sudol M, He JC, Mundel P: Yes-associated protein (YAP) promotes cell survival by inhibiting proapoptotic dendrin signaling. J Biol Chem 288: 17057–17062, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Boerries M, Grahammer F, Eiselein S, Buck M, Meyer C, Goedel M, Bechtel W, Zschiedrich S, Pfeifer D, Laloë D, Arrondel C, Gonçalves S, Krüger M, Harvey SJ, Busch H, Dengjel J, Huber TB: Molecular fingerprinting of the podocyte reveals novel gene and protein regulatory networks. Kidney Int 83: 1052–1064, 2013 [DOI] [PubMed] [Google Scholar]