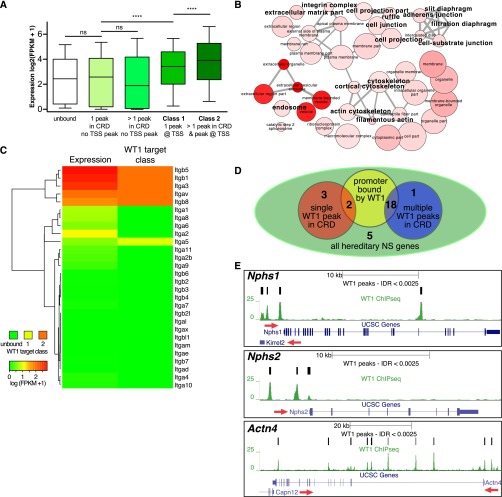
Figure 2.
WT1 activates a podocyte-specific transcriptome and targets most hereditary podocytopathy genes. (A) Box plots correlating WT1 target gene classes and binding patterns to podocyte gene expression levels by RNAseq. The presence of WT1 binding sites at target gene TSSs (class 1) is associated with higher absolute mRNA expression level average when compared with the average expression level of genes lacking a WT1 binding TSS. The average absolute expression level of genes positive for genic/intergenic WT1 peaks in addition to a WT1-bound TSS (class 2) further exceeds the average level of class 1 gene expression. The average expression level of genes positive for WT1 peaks within their cis-regulatory domains, but without a peak close to their TSSs, did not differ from unbound genes, suggesting that the predominant effect of WT1 in podocytes is transcription activation. ns, not significant. ****P<0.001. (B) GO enrichment network for cellular components derived from class 2 WT1 target genes shows enrichment of major podocyte components. Color intensity increases with significance, node size represents abundance of term in database, and edge strength indicates similarity. A larger font size indicates terms with particular relevance to podocytes. (C) Heat map and clustering analysis correlating expression levels of the integrin gene family to WT1 target classes. A class 2 binding pattern specifically identifies integrins expressed in podocytes. (D) Venn diagram summarizing binding patterns at hereditary podocytopathy genes. Eighteen genes are bound in a class 2 pattern, with a further two genes in a class 1 pattern. (E) Genome browser plots of WT1 binding sites, gene location, and conservation showing class 2 binding patterns at the three key hereditary NS/FSGS genes, Nphs1, Nphs2, and Actn4. Red arrows indicate direction of transcription.